A Column Level Lineage Graph for SQL.
Have you ever wondered what is the column level relationship among your SQL scripts and base tables? Don't worry, this tool is intended to help you by creating an interactive graph on a webpage to explore the column level lineage among them(Currently only supports Postgres, other connection types or dialects are under development).
Here is a live demo with the mimic-iv concepts_postgres files(navigation instructions) and that is created with one line of code:
from lineagex.lineagex import lineagex
lineagex(sql=path/to/sql, target_schema="schema1", conn_string="postgresql://username:password@server:port/database", search_path_schema="schema1, public")Check out more detailed usage and examples here.
The input can be a path to a SQL file, a path to a folder containing SQL files, a list of SQLs or a list of view names and/or schemas. Optionally, you can provide less information with only the SQLs, but providing the schema information and database connection is highly recommended for the best result.
The output would be a output.json and a index.html file in the folder. Start a local http server and you would be able to see the interactive graph.
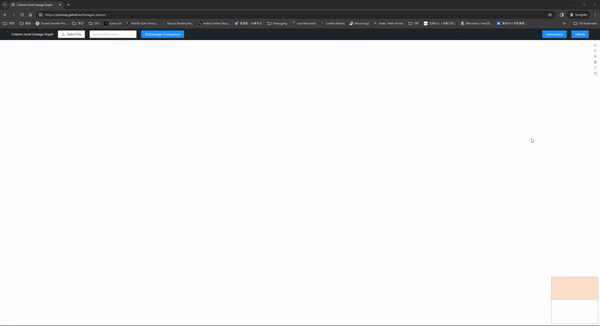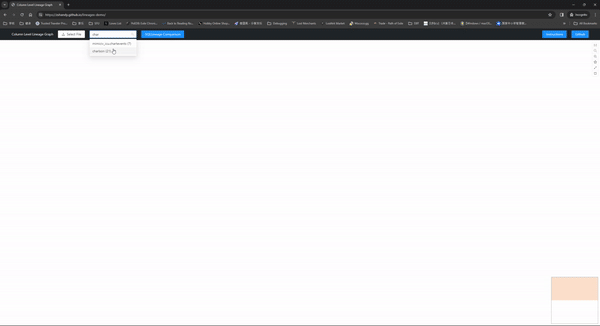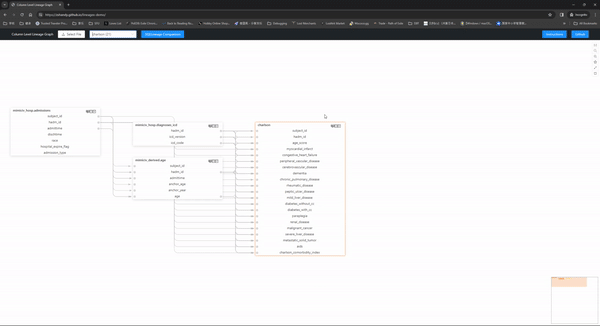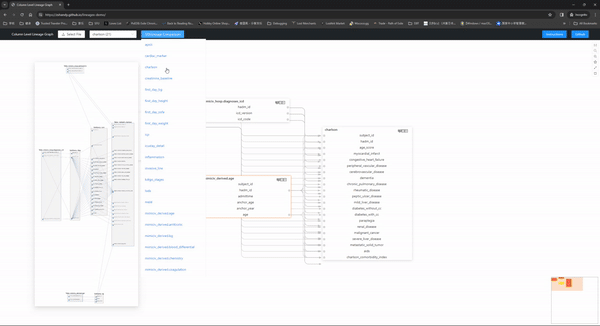 Check out more detailed navigation instructions here.
Check out more detailed navigation instructions here.
A general introduction of the project can be found in this blog post.
- Automatic dependency creation: When there are dependency among the SQL files, and those tables are not yet in the database, LineageX will automatically tries to find the dependency table and creates it.
- Clean and simple but very interactive user interface: The user interface is very simple to use with minimal clutters on the page while showing all of the necessary information.
- Variety of SQL statements: LineageX supports a variety of SQL statements, aside from the typical
SELECTstatement, it also supportsCREATE TABLE/VIEW [IF NOT EXISTS]statement as well as theINSERTandDELETEstatement. - dbt support: LineageX also implemented in the dbt-LineageX, it is added into a dbt project and by using the dbt library fal, it is able to reuse the Python core and create the similar output from the dbt project.
You can upload JSON files into the HTML produced and draw its lineage graph. Here is the supported format:
{
table_name: {
tables:[],
columns:{
column1: [[], []], // The first element is the list of columns that contribute directly to column1,
// The second element is the list of columns that are referenced, such as columns from WHERE/GROUP BY
column2: [[], []]
},
table_name: "",
sql: "",
},
}As an example:
{
table1: {
tables: [schema1.other_table],
columns: {
column1: [[schema1.other_table.columns1], [schema1.other_table.columns3]],
column2: [[schema1.other_table.columns2], [schema1.other_table.columns3]]
},
table_name: schema1.table1,
sql: SELECT column1, column2 FROM schema1.other_table WHERE column3 IS NOT NULL;
},
}When entering the conn_string parameter, only supported databases' connection types can be parsed successfully, or the lineage graph would be created as if no conn_string parameter is given.
- Postgres
- dbt-Postgres
- Mysql
- Sqlite
- SQL Server
- Oracle
- ...
Doc: https://sfu-db.github.io/lineagex/intro.html or just here