- Documentation: https://niaarm.readthedocs.io/en/latest
- Tested OS: Windows, Ubuntu, Fedora, Alpine, Arch, macOS. However, that does not mean it does not work on others
NiaARM is a framework for Association Rule Mining based on nature-inspired algorithms for optimization. 🌿 The framework is written fully in Python and runs on all platforms. NiaARM allows users to preprocess the data in a transaction database automatically, to search for association rules and provide a pretty output of the rules found. 📊 This framework also supports integral and real-valued types of attributes besides the categorical ones. Mining the association rules is defined as an optimization problem, and solved using the nature-inspired algorithms that come from the related framework called NiaPy. 🔗
The current version includes (but is not limited to) the following functions:
- loading datasets in CSV format 📁
- preprocessing of data 🧹
- searching for association rules 🔎
- providing output of mined association rules 📋
- generating statistics about mined association rules 📊
- visualization of association rules 📈
- association rule text mining (experimental) 📄
Install NiaARM with pip:
pip install niaarmTo install NiaARM on Alpine Linux, please enable Community repository and use:
$ apk add py3-niaarmTo install NiaARM on Arch Linux, please use an AUR helper:
$ yay -Syyu python-niaarmTo install NiaARM on Fedora, use:
$ dnf install python3-niaarmTo install NiaARM on NixOS, please use:
nix-env -iA nixos.python311Packages.niaarmIn NiaARM, data loading is done via the Dataset class. There are two options for loading data:
import pandas as pd
from niaarm import Dataset
df = pd.read_csv('datasets/Abalone.csv')
# preprocess data...
data = Dataset(df)
print(data) # printing the dataset will generate a feature reportfrom niaarm import Dataset
data = Dataset('datasets/Abalone.csv')
print(data)Optionally, a preprocessing technique, called data squashing [5], can be applied. This will significantly reduce the number of transactions, while providing similar results to the original dataset.
from niaarm import Dataset, squash
dataset = Dataset('datasets/Abalone.csv')
squashed = squash(dataset, threshold=0.9, similarity='euclidean')
print(squashed)Association rule mining can be easily performed using the get_rules function:
from niaarm import Dataset, get_rules
from niapy.algorithms.basic import DifferentialEvolution
data = Dataset("datasets/Abalone.csv")
algo = DifferentialEvolution(population_size=50, differential_weight=0.5, crossover_probability=0.9)
metrics = ('support', 'confidence')
rules, run_time = get_rules(data, algo, metrics, max_iters=30, logging=True)
print(rules) # Prints basic stats about the mined rules
print(f'Run Time: {run_time}')
rules.to_csv('output.csv')The above example can be also be implemented using a more low level interface,
with the NiaARM class directly:
from niaarm import NiaARM, Dataset
from niapy.algorithms.basic import DifferentialEvolution
from niapy.task import Task, OptimizationType
data = Dataset("datasets/Abalone.csv")
# Create a problem
# dimension represents the dimension of the problem;
# features represent the list of features, while transactions depicts the list of transactions
# metrics is a sequence of metrics to be taken into account when computing the fitness;
# you can also pass in a dict of the shape {'metric_name': <weight of metric in range [0, 1]>};
# when passing a sequence, the weights default to 1.
problem = NiaARM(data.dimension, data.features, data.transactions, metrics=('support', 'confidence'), logging=True)
# build niapy task
task = Task(problem=problem, max_iters=30, optimization_type=OptimizationType.MAXIMIZATION)
# use Differential Evolution (DE) algorithm from the NiaPy library
# see full list of available algorithms: https://github.com/NiaOrg/NiaPy/blob/master/Algorithms.md
algo = DifferentialEvolution(population_size=50, differential_weight=0.5, crossover_probability=0.9)
# run algorithm
best = algo.run(task=task)
# sort rules
problem.rules.sort()
# export all rules to csv
problem.rules.to_csv('output.csv')The framework implements several popular interest measures, which can be used to compute the fitness function value of rules and for assessing the quality of the mined rules. A full list of the implemented interest measures along with their descriptions and equations can be found here.
The framework currently supports:
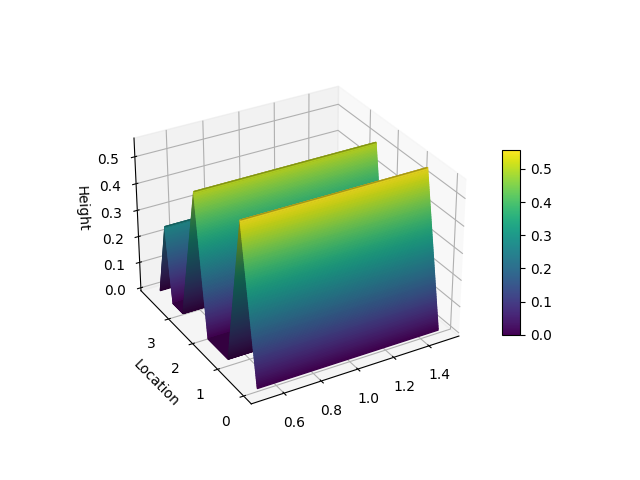
- hill slopes (presented in [4]),
- scatter plot and
- grouped matrix plot visualization methods.
More visualization methods are planned to be implemented in future releases.
from matplotlib import pyplot as plt
from niaarm import Dataset, get_rules
from niaarm.visualize import hill_slopes
dataset = Dataset('datasets/Abalone.csv')
metrics = ('support', 'confidence')
rules, _ = get_rules(dataset, 'DifferentialEvolution', metrics, max_evals=1000, seed=1234)
some_rule = rules[150]
hill_slopes(some_rule, dataset.transactions)
plt.show()from examples.visualization_examples.prepare_datasets import get_weather_data
from niaarm import Dataset, get_rules
from niaarm.visualize import scatter_plot
# Get prepared data
arm_df = get_weather_data()
# Prepare Dataset
dataset = Dataset(path_or_df=arm_df,delimiter=",")
# Get rules
metrics = ("support", "confidence")
rules, run_time = get_rules(dataset, "DifferentialEvolution", metrics, max_evals=500)
# Add lift to metrics
metrics = list(metrics)
metrics.append("lift")
metrics = tuple(metrics)
# Visualize scatter plot
fig = scatter_plot(rules=rules, metrics=metrics, interactive=False)
fig.show()from examples.visualization_examples.prepare_datasets import get_football_player_data
from niaarm import Dataset, get_rules
from niaarm.visualize import grouped_matrix_plot
# Get prepared data
arm_df = get_football_player_data()
# Prepare Dataset
dataset = Dataset(path_or_df=arm_df, delimiter=",")
# Get rules
metrics = ("support", "confidence")
rules, run_time = get_rules(dataset, "DifferentialEvolution", metrics, max_evals=500)
# Add lift to metrics
metrics = list(metrics)
metrics.append("lift")
metrics = tuple(metrics)
# Visualize grouped matrix plot
fig = grouped_matrix_plot(rules=rules, metrics=metrics, k=5, interactive=False)
fig.show()An experimental implementation of association rule text mining using nature-inspired algorithms, based on ideas from [5]
is also provided. The niaarm.text module contains the Corpus and Document classes for loading and preprocessing corpora,
a TextRule class, representing a text rule, and the NiaARTM class, implementing association rule text mining
as a continuous optimization problem. The get_text_rules function, equivalent to get_rules, but for text mining, was also
added to the niaarm.mine module.
import pandas as pd
from niaarm.text import Corpus
from niaarm.mine import get_text_rules
from niapy.algorithms.basic import ParticleSwarmOptimization
df = pd.read_json('datasets/text/artm_test_dataset.json', orient='records')
documents = df['text'].tolist()
corpus = Corpus.from_list(documents)
algorithm = ParticleSwarmOptimization(population_size=200, seed=123)
metrics = ('support', 'confidence', 'aws')
rules, time = get_text_rules(corpus, max_terms=5, algorithm=algorithm, metrics=metrics, max_evals=10000, logging=True)
print(rules)
print(f'Run time: {time:.2f}s')
rules.to_csv('output.csv')Note: You may need to download stopwords and the punkt tokenizer from nltk by running import nltk; nltk.download('stopwords'); nltk.download('punkt').
For a full list of examples see the examples folder in the GitHub repository.
We provide a simple command line interface, which allows you to easily mine association rules on any input dataset, output them to a csv file and/or perform a simple statistical analysis on them. For more details see the documentation.
niaarm -husage: niaarm [-h] [-v] [-c CONFIG] [-i INPUT_FILE] [-o OUTPUT_FILE] [--squashing-similarity {euclidean,cosine}] [--squashing-threshold SQUASHING_THRESHOLD] [-a ALGORITHM] [-s SEED] [--max-evals MAX_EVALS] [--max-iters MAX_ITERS]
[--metrics METRICS [METRICS ...]] [--weights WEIGHTS [WEIGHTS ...]] [--log] [--stats]
Perform ARM, output mined rules as csv, get mined rules' statistics
options:
-h, --help show this help message and exit
-v, --version show program's version number and exit
-c CONFIG, --config CONFIG
Path to a TOML config file
-i INPUT_FILE, --input-file INPUT_FILE
Input file containing a csv dataset
-o OUTPUT_FILE, --output-file OUTPUT_FILE
Output file for mined rules
--squashing-similarity {euclidean,cosine}
Similarity measure to use for squashing
--squashing-threshold SQUASHING_THRESHOLD
Threshold to use for squashing
-a ALGORITHM, --algorithm ALGORITHM
Algorithm to use (niapy class name, e.g. DifferentialEvolution)
-s SEED, --seed SEED Seed for the algorithm's random number generator
--max-evals MAX_EVALS
Maximum number of fitness function evaluations
--max-iters MAX_ITERS
Maximum number of iterations
--metrics METRICS [METRICS ...]
Metrics to use in the fitness function.
--weights WEIGHTS [WEIGHTS ...]
Weights in range [0, 1] corresponding to --metrics
--log Enable logging of fitness improvements
--stats Display stats about mined rules
Note: The CLI script can also run as a python module (python -m niaarm ...)
Ideas are based on the following research papers:
[1] I. Fister Jr., A. Iglesias, A. Gálvez, J. Del Ser, E. Osaba, I Fister. Differential evolution for association rule mining using categorical and numerical attributes In: Intelligent data engineering and automated learning - IDEAL 2018, pp. 79-88, 2018.
[2] I. Fister Jr., V. Podgorelec, I. Fister. Improved Nature-Inspired Algorithms for Numeric Association Rule Mining. In: Vasant P., Zelinka I., Weber GW. (eds) Intelligent Computing and Optimization. ICO 2020. Advances in Intelligent Systems and Computing, vol 1324. Springer, Cham.
[3] I. Fister Jr., I. Fister A brief overview of swarm intelligence-based algorithms for numerical association rule mining. arXiv preprint arXiv:2010.15524 (2020).
[4] Fister, I. et al. (2020). Visualization of Numerical Association Rules by Hill Slopes. In: Analide, C., Novais, P., Camacho, D., Yin, H. (eds) Intelligent Data Engineering and Automated Learning – IDEAL 2020. IDEAL 2020. Lecture Notes in Computer Science(), vol 12489. Springer, Cham. https://doi.org/10.1007/978-3-030-62362-3_10
[5] I. Fister, S. Deb, I. Fister, Population-based metaheuristics for Association Rule Text Mining, In: Proceedings of the 2020 4th International Conference on Intelligent Systems, Metaheuristics & Swarm Intelligence, New York, NY, USA, mar. 2020, pp. 19–23. doi: 10.1145/3396474.3396493.
[6] I. Fister, I. Fister Jr., D. Novak and D. Verber, Data squashing as preprocessing in association rule mining, 2022 IEEE Symposium Series on Computational Intelligence (SSCI), Singapore, Singapore, 2022, pp. 1720-1725, doi: 10.1109/SSCI51031.2022.10022240.
[1] NiaARM.jl: Numerical Association Rule Mining in Julia
This package is distributed under the MIT License. This license can be found online at http://www.opensource.org/licenses/MIT.
This framework is provided as-is, and there are no guarantees that it fits your purposes or that it is bug-free. Use it at your own risk!
Stupan, Ž., & Fister Jr., I. (2022). NiaARM: A minimalistic framework for Numerical Association Rule Mining. Journal of Open Source Software, 7(77), 4448.
Thanks goes to these wonderful people (emoji key):
zStupan 💻 🐛 📖 🖋 🤔 💡 |
Iztok Fister Jr. 💻 🐛 🧑🏫 🚧 🤔 |
Erkan Karabulut 💻 🐛 |
Tadej Lahovnik 📖 |
Ben Beasley 📖 |
Dusan Fister 🎨 |
This project follows the all-contributors specification. Contributions of any kind welcome!