diff --git a/README.md b/README.md
index d65ddc70..cb9be8ff 100644
--- a/README.md
+++ b/README.md
@@ -9,9 +9,11 @@ R package for Airborne LiDAR Data Manipulation and Visualization for Forestry Ap
The lidR package provides functions to read and write `.las` and `.laz` files, plot point clouds, compute metrics using an area-based approach, compute digital canopy models, thin LiDAR data, manage a collection of LAS/LAZ files, automatically extract ground inventories, process a collection of tiles using multicore processing, segment individual trees, classify points from geographic data, and provides other tools to manipulate LiDAR data in a research and development context.
-:book: Read [the book](https://r-lidar.github.io/lidRbook/index.html) to get started with the lidR package. See changelogs on [NEW.md](https://github.com/r-lidar/lidR/blob/master/NEWS.md)
+- 📖 Read [the book](https://r-lidar.github.io/lidRbook/index.html) to get started with the lidR package. See changelogs on [NEW.md](https://github.com/r-lidar/lidR/blob/master/NEWS.md)
+- 💻 Install `lidR` from R with: `install.packages("lidR")`
+- 💵 [Sponsor `lidR`](https://github.com/sponsors/Jean-Romain)
-To cite the package use `citation()` from within R:
+`lidR` has been cited by more than 1,000 scientific papers. To cite the package use `citation()` from within R:
```r
citation("lidR")
@@ -49,6 +51,8 @@ chm <- rasterize_canopy(las, 1, pitfree(thr, edg))
plot(chm)
```
+
+
### Read and display a catalog of las files
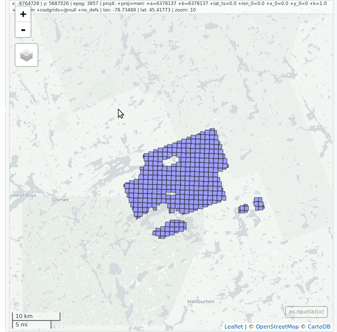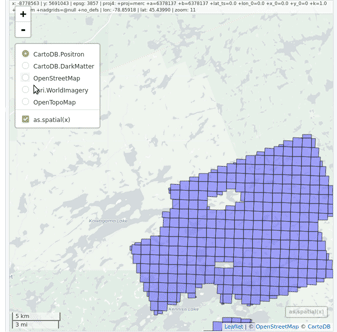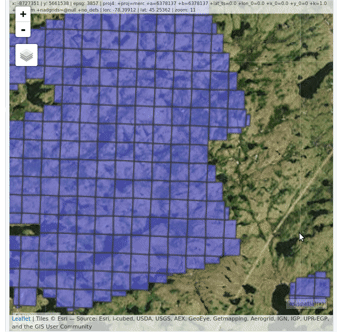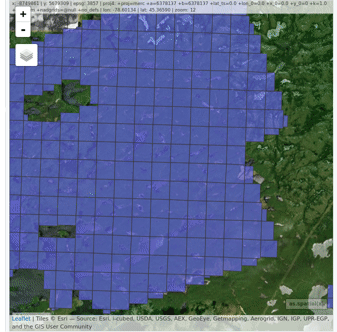 @@ -62,9 +66,11 @@ plot(ctg, map = TRUE)
From a `LAScatalog` object the user can (for example) extract some regions of interest (ROI) with `clip_roi()`. Using a catalog for the extraction of the ROI guarantees fast and memory-efficient clipping. `LAScatalog` objects allow many other manipulations that can be done with multicore processing.
+
@@ -62,9 +66,11 @@ plot(ctg, map = TRUE)
From a `LAScatalog` object the user can (for example) extract some regions of interest (ROI) with `clip_roi()`. Using a catalog for the extraction of the ROI guarantees fast and memory-efficient clipping. `LAScatalog` objects allow many other manipulations that can be done with multicore processing.
+
+
### Individual tree segmentation
-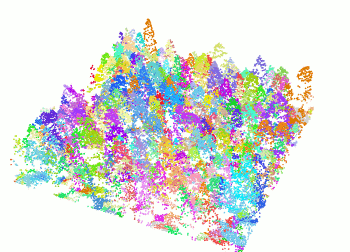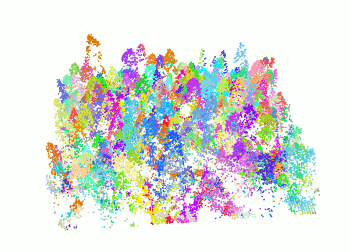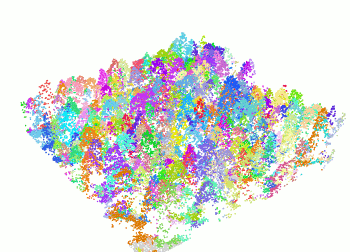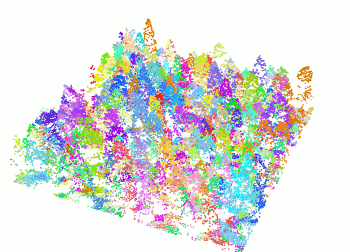 +
+ The `segment_trees()` function has several algorithms from the literature for individual tree segmentation, based either on the digital canopy model or on the point-cloud. Each algorithm has been coded from the source article to be as close as possible to what was written in the peer-reviewed papers. Our goal is to make published algorithms usable, testable and comparable.
@@ -111,10 +117,12 @@ plot(las, color = "Amplitude", add = x)
# About
-**lidR** is developed openly at [Laval University](https://www.ulaval.ca/en).
+**lidR** is developed openly by [r-lidar](https://www.r-lidar.com/).
-* Development of the `lidR` package between 2015 and 2018 was made possible thanks to the financial support of the [AWARE project (NSERC CRDPJ 462973-14)](https://awareproject.ca/); grantee [Prof Nicholas Coops](https://forestry.ubc.ca/faculty-profile/nicholas-coops/).
-* Development of the `lidR` package between 2018 and 2021 was made possible thanks to the financial support of the [Ministère des Ressources naturelles et des Forêts](https://www.quebec.ca/en/government/ministere/ressources-naturelles-forets).
+The development of the `lidR` package was made possible in the past thanks to the financial support of:
+* 2015-2018: the [AWARE project (NSERC CRDPJ 462973-14)](https://awareproject.ca/); grantee [Prof Nicholas Coops](https://forestry.ubc.ca/faculty-profile/nicholas-coops/).
+* 2018-2021: the [Ministère des Ressources naturelles et des Forêts](https://www.quebec.ca/en/government/ministere/ressources-naturelles-forets).
+* 2021-2024: the [Laval University](https://www.ulaval.ca/en).
The `segment_trees()` function has several algorithms from the literature for individual tree segmentation, based either on the digital canopy model or on the point-cloud. Each algorithm has been coded from the source article to be as close as possible to what was written in the peer-reviewed papers. Our goal is to make published algorithms usable, testable and comparable.
@@ -111,10 +117,12 @@ plot(las, color = "Amplitude", add = x)
# About
-**lidR** is developed openly at [Laval University](https://www.ulaval.ca/en).
+**lidR** is developed openly by [r-lidar](https://www.r-lidar.com/).
-* Development of the `lidR` package between 2015 and 2018 was made possible thanks to the financial support of the [AWARE project (NSERC CRDPJ 462973-14)](https://awareproject.ca/); grantee [Prof Nicholas Coops](https://forestry.ubc.ca/faculty-profile/nicholas-coops/).
-* Development of the `lidR` package between 2018 and 2021 was made possible thanks to the financial support of the [Ministère des Ressources naturelles et des Forêts](https://www.quebec.ca/en/government/ministere/ressources-naturelles-forets).
+The development of the `lidR` package was made possible in the past thanks to the financial support of:
+* 2015-2018: the [AWARE project (NSERC CRDPJ 462973-14)](https://awareproject.ca/); grantee [Prof Nicholas Coops](https://forestry.ubc.ca/faculty-profile/nicholas-coops/).
+* 2018-2021: the [Ministère des Ressources naturelles et des Forêts](https://www.quebec.ca/en/government/ministere/ressources-naturelles-forets).
+* 2021-2024: the [Laval University](https://www.ulaval.ca/en).

 @@ -62,9 +66,11 @@ plot(ctg, map = TRUE)
From a `LAScatalog` object the user can (for example) extract some regions of interest (ROI) with `clip_roi()`. Using a catalog for the extraction of the ROI guarantees fast and memory-efficient clipping. `LAScatalog` objects allow many other manipulations that can be done with multicore processing.
+
@@ -62,9 +66,11 @@ plot(ctg, map = TRUE)
From a `LAScatalog` object the user can (for example) extract some regions of interest (ROI) with `clip_roi()`. Using a catalog for the extraction of the ROI guarantees fast and memory-efficient clipping. `LAScatalog` objects allow many other manipulations that can be done with multicore processing.
+ +
+ The `segment_trees()` function has several algorithms from the literature for individual tree segmentation, based either on the digital canopy model or on the point-cloud. Each algorithm has been coded from the source article to be as close as possible to what was written in the peer-reviewed papers. Our goal is to make published algorithms usable, testable and comparable.
@@ -111,10 +117,12 @@ plot(las, color = "Amplitude", add = x)
# About
-**lidR** is developed openly at [Laval University](https://www.ulaval.ca/en).
+**lidR** is developed openly by [r-lidar](https://www.r-lidar.com/).
-* Development of the `lidR` package between 2015 and 2018 was made possible thanks to the financial support of the [AWARE project (NSERC CRDPJ 462973-14)](https://awareproject.ca/); grantee [Prof Nicholas Coops](https://forestry.ubc.ca/faculty-profile/nicholas-coops/).
-* Development of the `lidR` package between 2018 and 2021 was made possible thanks to the financial support of the [Ministère des Ressources naturelles et des Forêts](https://www.quebec.ca/en/government/ministere/ressources-naturelles-forets).
+The development of the `lidR` package was made possible in the past thanks to the financial support of:
+* 2015-2018: the [AWARE project (NSERC CRDPJ 462973-14)](https://awareproject.ca/); grantee [Prof Nicholas Coops](https://forestry.ubc.ca/faculty-profile/nicholas-coops/).
+* 2018-2021: the [Ministère des Ressources naturelles et des Forêts](https://www.quebec.ca/en/government/ministere/ressources-naturelles-forets).
+* 2021-2024: the [Laval University](https://www.ulaval.ca/en).
The `segment_trees()` function has several algorithms from the literature for individual tree segmentation, based either on the digital canopy model or on the point-cloud. Each algorithm has been coded from the source article to be as close as possible to what was written in the peer-reviewed papers. Our goal is to make published algorithms usable, testable and comparable.
@@ -111,10 +117,12 @@ plot(las, color = "Amplitude", add = x)
# About
-**lidR** is developed openly at [Laval University](https://www.ulaval.ca/en).
+**lidR** is developed openly by [r-lidar](https://www.r-lidar.com/).
-* Development of the `lidR` package between 2015 and 2018 was made possible thanks to the financial support of the [AWARE project (NSERC CRDPJ 462973-14)](https://awareproject.ca/); grantee [Prof Nicholas Coops](https://forestry.ubc.ca/faculty-profile/nicholas-coops/).
-* Development of the `lidR` package between 2018 and 2021 was made possible thanks to the financial support of the [Ministère des Ressources naturelles et des Forêts](https://www.quebec.ca/en/government/ministere/ressources-naturelles-forets).
+The development of the `lidR` package was made possible in the past thanks to the financial support of:
+* 2015-2018: the [AWARE project (NSERC CRDPJ 462973-14)](https://awareproject.ca/); grantee [Prof Nicholas Coops](https://forestry.ubc.ca/faculty-profile/nicholas-coops/).
+* 2018-2021: the [Ministère des Ressources naturelles et des Forêts](https://www.quebec.ca/en/government/ministere/ressources-naturelles-forets).
+* 2021-2024: the [Laval University](https://www.ulaval.ca/en).