@@ -154,8 +155,8 @@ For this example we assume an image with 5 dimensions and axes called `t,c,z,y,x
├── n # The name of the array is arbitrary with the ordering defined by
│ │ # by the "multiscales" metadata, but is often a sequence starting at 0.
│ │
- │ ├── .zarray # All image arrays must be up to 5-dimensional
- │ │ # with the axis of type time before type channel, before spatial axes.
+ │ ├── .zarray
+ │ │
│ │
│ └─ t # Chunks are stored with the nested directory layout.
│ └─ c # All but the last chunk element are stored as directories.
@@ -168,7 +169,7 @@ For this example we assume an image with 5 dimensions and axes called `t,c,z,y,x
├── .zgroup # The labels group is a container which holds a list of labels to make the objects easily discoverable
│
├── .zattrs # All labels will be listed in `.zattrs` e.g. `{ "labels": [ "original/0" ] }`
- │ # Each dimension of the label `(t, c, z, y, x)` should be either the same as the
+ │ # Each dimension of the label should be either the same as the
│ # corresponding dimension of the image, or `1` if that dimension of the label
│ # is irrelevant.
│
@@ -189,7 +190,7 @@ High-content screening {#hcs-layout}
------------------------------------
The following specification defines the hierarchy for a high-content screening
-dataset. Three groups MUST be defined above the images:
+dataset. Three groups must be defined above the images:
- the group above the images defines the well and MUST implement the
[well specification](#well-md). All images contained in a well are fields
@@ -199,9 +200,6 @@ dataset. Three groups MUST be defined above the images:
collection of wells organized in rows and columns. It MUST implement the
[plate specification](#plate-md)
-A well row group SHOULD NOT be present if there are no images in the well row.
-A well group SHOULD NOT be present if there are no images in the well.
-
. # Root folder, potentially in S3,
@@ -238,18 +236,6 @@ Metadata {#metadata}
The various `.zattrs` files throughout the above array hierarchy may contain metadata
keys as specified below for discovering certain types of data, especially images.
-"axes" metadata {#axes-md}
---------------------------
-
-"axes" describes the dimensions of a physical coordinate space. It is a list of dictionaries, where each dictionary describes a dimension (axis) and:
-- MUST contain the field "name" that gives the name for this dimension. The values MUST be unique across all "name" fields.
-- SHOULD contain the field "type". It SHOULD be one of "space", "time" or "channel", but MAY take other values for custom axis types that are not part of this specification yet.
-- SHOULD contain the field "unit" to specify the physical unit of this dimension. The value SHOULD be one of the following strings, which are valid units according to UDUNITS-2.
- - Units for "space" axes: 'angstrom', 'attometer', 'centimeter', 'decimeter', 'exameter', 'femtometer', 'foot', 'gigameter', 'hectometer', 'inch', 'kilometer', 'megameter', 'meter', 'micrometer', 'mile', 'millimeter', 'nanometer', 'parsec', 'petameter', 'picometer', 'terameter', 'yard', 'yoctometer', 'yottameter', 'zeptometer', 'zettameter'
- - Units for "time" axes: 'attosecond', 'centisecond', 'day', 'decisecond', 'exasecond', 'femtosecond', 'gigasecond', 'hectosecond', 'hour', 'kilosecond', 'megasecond', 'microsecond', 'millisecond', 'minute', 'nanosecond', 'petasecond', 'picosecond', 'second', 'terasecond', 'yoctosecond', 'yottasecond', 'zeptosecond', 'zettasecond'
-
-If part of [[#multiscale-md]], the length of "axes" MUST be equal to the number of dimensions of the arrays that contain the image data.
-
"bioformats2raw.layout" (transitional) {#bf2raw}
------------------------------------------------
@@ -331,44 +317,1106 @@ Conforming readers:
- MAY ignore other groups or arrays under the root of the hierarchy.
+"coordinateSystems" metadata {#coord-sys-md}
+--------------------------
+
+A "coordinate system" is a collection of "axes" / dimensions with a name. Every coordinate system:
+- MUST contain the field "name". The value MUST be a non-empty string that is unique among `coordinateSystem`s.
+- MUST contain the field "axes", whose value is an array of valid "axes" (see below).
+
+
+
+```json
+{
+ "name" : "volume_micrometers",
+ "axes" : [
+ {"name": "z", "type": "space", "unit": "micrometer"},
+ {"name": "y", "type": "space", "unit": "micrometer"},
+ {"name": "x", "type": "space", "unit": "micrometer"}
+ ]
+}
+```
+
+
+The order of the `"axes"` list matters and defines the index of each array dimension and coordinates for points in that
+coordinate system. For the above example, the `"x"` dimension is the first dimension. The "dimensionality" of a coordinate system
+is indicated by the length of its "axes" array. The "volume_micrometers" example coordinate system above is three dimensional (3D).
+
+The axes of a coordinate system (see below) give information about the types, units, and other properties of the coordinate
+system's dimensions. Axis `name`s may contain semantically meaningful information, but can be arbitrary. As a result, two
+coordinate systems that have identical axes in the same order may not be "the same" in the sense that measurements at the same
+point refer to different physical entities and therefore should not be analyzed jointly. Tasks that require images, annotations,
+regions of interest, etc., SHOULD ensure that they are in the same coordinate system (same name, with identical axes) or can be
+transformed to the same coordinate system before doing analysis. See the example below.
+
+
+
+Two instruments simultaneously image the same sample from two different angles, and the 3D data from both instruments are
+calibrated to "micrometer" units. Two samples are collected ("sampleA" and "sampleB"). An analysis of sample A requires
+measurements from both instruments' images at certain points in space. Suppose a region of interest (ROI) is determined from the
+image obtained from instrument 2, but quantification from that region is needed for instrument 1. Since measurements were
+collected at different angles, a measurement by instrument 1 at the point with coordinates (x,y,z) may not correspond to the
+measurement at the same point in instrument 2 (i.e., it may not be the same physical location in the sample). To analyze both
+images together, they must be in the same coordinate system.
+
+The set of coordinate transformations ([[#trafo-md]]) encodes relationships between coordinate systems, specifically, how to
+convert points and images to different coordinate systems. Implementations can apply the coordinate transform to images or
+points in coordinate system "sampleA_instrument2" to bring them into the "sampleA_instrument1" coordinate system. In this case,
+the ROI should be transformed to the "sampleA_image1" coordinate system, then used for quantification with the instrument 1
+image.
+
+```json
+"coordinateSystems" : [
+ {
+ "name" : "sampleA-instrument1",
+ "axes" : [
+ {"name": "z", "type": "space", "unit": "micrometer"},
+ {"name": "y", "type": "space", "unit": "micrometer"},
+ {"name": "x", "type": "space", "unit": "micrometer"}
+ ]
+ },
+ {
+ "name" : "sampleA-instrument2",
+ "axes" : [
+ {"name": "z", "type": "space", "unit": "micrometer"},
+ {"name": "y", "type": "space", "unit": "micrometer"},
+ {"name": "x", "type": "space", "unit": "micrometer"}
+ ]
+ }
+],
+"coordinateTransformations": [
+ {
+ "type": "affine":
+ "path": "../sampleA_instrument2-to-instrument1"
+ "input": "sampleA_instrument2",
+ "output": "sampleA_instrument1"
+ }
+]
+```
+
+
+
+
+
+### "axes" metadata
+
+"axes" describes the dimensions of a coordinate systems. It is a list of dictionaries, where each dictionary describes a dimension (axis) and:
+- MUST contain the field "name" that gives the name for this dimension. The values MUST be unique across all "name" fields.
+- SHOULD contain the field "type". It SHOULD be one of the strings "array", "space", "time", "channel", "coordinate", or
+ "displacement" but MAY take other string values for custom axis types that are not part of this specification yet.
+- MAY contain the field "discrete". The value MUST be a boolean, and is `true` if the axis represents a discrete dimension.
+- SHOULD contain the field "unit" to specify the physical unit of this dimension. The value SHOULD be one of the following strings, which are valid units according to UDUNITS-2.
+ - Units for "space" axes: 'angstrom', 'attometer', 'centimeter', 'decimeter', 'exameter', 'femtometer', 'foot', 'gigameter', 'hectometer', 'inch', 'kilometer', 'megameter', 'meter', 'micrometer', 'mile', 'millimeter', 'nanometer', 'parsec', 'petameter', 'picometer', 'terameter', 'yard', 'yoctometer', 'yottameter', 'zeptometer', 'zettameter'
+ - Units for "time" axes: 'attosecond', 'centisecond', 'day', 'decisecond', 'exasecond', 'femtosecond', 'gigasecond', 'hectosecond', 'hour', 'kilosecond', 'megasecond', 'microsecond', 'millisecond', 'minute', 'nanosecond', 'petasecond', 'picosecond', 'second', 'terasecond', 'yoctosecond', 'yottasecond', 'zeptosecond', 'zettasecond'
+- MAY contain the field "longName". The value MUST be a string, and can provide a longer name or description of an axis and its properties.
+
+If part of [[#multiscale-md]], the length of "axes" MUST be equal to the number of dimensions of the arrays that contain the image data.
+
+
+
+Examples of valid axes:
+
+```json
+[
+ {"name": "x", "type": "space", "unit": "micrometer"},
+ {"name": "t", "type": "time", "unit": "second", "longName": "Unix Epoch time"},
+ {"name": "c", "type": "channel", "discrete": true},
+ {"name": "i0", "type": "array"},
+ {"name": "c", "type": "coordinate", "discrete" : true },
+ {"name": "v", "type": "displacement", "discrete": true },
+ {"name": "freq", "type": "frequency", "unit": "megahertz"}
+]
+```
+
+
+Arrays are inherently discrete (see Array coordinate systems, below) but are often used to store discrete samples of a
+continuous variable. The continuous values "in between" discrete samples can be retrieved using an *interpolation* method. If an
+axis is continuous (`"discrete" : false`), it indicates that interpolation is well-defined. Axes representing `space` and
+`time` are usually continuous. Similarly, joint interpolation across axes is well-defined only for axes of the same `type`. In
+contrast, discrete axes (`"discrete" : true`) may be indexed only by integers. Axes of representing a `channel`, `coordinate`,
+or `displacement` are usually discrete.
+
+Note: The most common methods for interpolation are "nearest neighbor", "linear", "cubic", and "windowed sinc". Here, we refer
+to any method that obtains values at real valued coordinates using discrete samples as an "interpolator". As such, label images
+may be interpolated using "nearest neighbor" to obtain labels at points along the continuum.
+
+
+
+For the coordinate system:
+
+```json
+{
+ "name" : "index and interpolation",
+ "axes" : [
+ {"name": "t", "type": "time"},
+ {"name": "c", "type": "channel", "discrete": true},
+ {"name": "y", "type": "space"},
+ {"name": "x", "type": "space"}
+ ]
+}
+```
+
+Indexing an image at the point `(0.1, 0.2, 0.3, 0.4)` is not valid, because the value of the first coordinate (`0.1`) refers
+to the discrete axis `"c"`. Indexing an image at the point `(1, 0.2, 0.3, 0.4)` is valid.
+
+
+
+### Array coordinate systems
+
+Every array has a default coordinate system whose parameters need not be explicitly defined. Its name is the path to the array
+in the container, its axes have `"type":"array"`, are unitless, and have default "name"s. The ith axis has `"name":"dim_i"`
+(these are the same default names used by [xarray](https://docs.xarray.dev/en/stable/user-guide/terminology.html)).
+
+For example, a 3D array at path `my/data/array` defines the coordinate system:
+
+```json
+{
+ "name" : "my/data/array",
+ "axes" : [
+ {"name": "dim_0", "type": "array"},
+ {"name": "dim_1", "type": "array"},
+ {"name": "dim_2", "type": "array"}
+ ]
+}
+```
+
+though this object should not and need not explicitly appear in metadata.
+
+
+
+The dimensionality of each array coordinate system equals the dimensionality of its corresponding zarr array. The axis with
+name `"dim_i"` is the ith element of the `"axes"` list. The axes and their order align with the `shape`
+attribute in the zarr array attributes (in `.zarray`), and whose data depends on the byte order used to store
+chunks. As described in the [zarr array metadata](https://zarr.readthedocs.io/en/stable/spec/v2.html#arrays),
+the last dimension of an array in "C" order are stored contiguously on disk or in-memory when directly loaded.
+
+
+For example, if `my/data/array/.zarray` contains:
+
+```json
+{
+ "chunks": [ 4, 3, 5 ],
+ "compressor": null,
+ "dtype": "|u1",
+ "fill_value": 0,
+ "filters": null,
+ "order": "C",
+ "shape": [ 4, 3, 5 ],
+ "zarr_format": 2
+}
+```
+
+Then `dim_0` has length 4, `dim_1` has length 3, and `dim_2` has length 5.
+
+
+The name and axes names MAY be customized by including a `arrayCoordinateSystem` field in
+the user-defined attributes of the array whose value is a coordinate system object. The length of
+`axes` MUST be equal to the dimensionality. The value of `"type"` for each object in the
+axes array MUST equal `"array"`.
+
+
+
+
+path: examples/coordSystems/arrayCoordSys.json
+highlight: json
+
+
+Note that dimension `i` is contiguous in memory.
+
+
+
+
+### Coordinate convention
+
+**The pixel/voxel center is the origin of the continuous coordinate system.**
+
+It is vital to consistently define relationship between the discrete/array and continuous/interpolated
+coordinate systems. A pixel/voxel is the continuous region (rectangle) that corresponds to a single sample
+in the discrete array, i.e., the area corresponding to nearest-neighbor (NN) interpolation of that sample.
+The center of a 2d pixel corresponding to the origin `(0,0)` in the discrete array is the origin of the continuous coordinate
+system `(0.0, 0.0)` (when the transformation is the identity). The continuous rectangle of the pixel is given by the
+half-open interval `[-0.5, 0.5) x [-0.5, 0.5)` (i.e., -0.5 is included, +0.5 is excluded). See chapter 4 and figure 4.1 of the ITK Software Guide [[itk]].
+
+
"coordinateTransformations" metadata {#trafo-md}
------------------------------------------------
-"coordinateTransformations" describe a series of transformations that map between two coordinate spaces (defined by "axes").
-For example, to map a discrete data space of an array to the corresponding physical space.
-It is a list of dictionaries. Each entry describes a single transformation and MUST contain the field "type".
-The value of "type" MUST be one of the elements of the `type` column in the table below.
-Additional fields for the entry depend on "type" and are defined by the column `fields`.
+"coordinateTransformations" describe the mapping between two coordinate systems (defined by "axes").
+For example, to map an array's discrete coordinate system to its corresponding physical coordinates.
+Coordinate transforms are in the "forward" direction. They represent functions from *points* in the
+input space to *points* in the output space.
+
+
+- MUST contain the field "type".
+- MUST contain any other fields required by the given "type" (see table below).
+- MUST contain the field "output", unless part of a `sequence` or `inverseOf` (see details).
+- MUST contain the field "input", unless part of a `sequence` or `inverseOf` (see details).
+- MAY contain the field "name". Its value MUST be unique across all "name" fields for coordinate transformations.
+- Parameter values MUST be compatible with input and output space dimensionality (see details).
| `identity` | identity transformation, is the default transformation and is typically not explicitly defined - | ||||
|---|---|---|---|---|---|
| `translation` | one of: `"translation":List[float]`, `"path":str` | translation vector, stored either as a list of floats (`"translation"`) or as binary data at a location in this container (`path`). The length of vector defines number of dimensions. | - | |||
| `scale` | one of: `"scale":List[float]`, `"path":str` | scale vector, stored either as a list of floats (`scale`) or as binary data at a location in this container (`path`). The length of vector defines number of dimensions. | + | |||
| `identity` + | + | The identity transformation is the default transformation and is typically not explicitly defined. + | |||
| `mapAxis` + | `"mapAxis":Dict[String:String]` + | A `maxAxis` transformation specifies an axis permutation as a map between axis names. + | |||
| `translation` + | one of: `"translation":List[number]`, `"path":str` + | translation vector, stored either as a list of numbers (`"translation"`) or as binary data at a location + in this container (`path`). + | |||
| `scale` + | one of: `"scale":List[number]`, `"path":str` + | scale vector, stored either as a list of numbers (`scale`) or as binary data at a location in this + container (`path`). + | |||
| `affine` + | one of: `"affine":List[List[number]]`, `"path":str` + | affine transformation matrix stored as a flat array stored either with json uing the affine field + or as binary data at a location in this container (path). If both are present, the binary values at path should be used. + | |||
| `rotation` + | one of: `"rotation":List[number]`, `"path":str` + | rotation transformation matrix stored as an array stored either + with json or as binary data at a location in this container (path). + If both are present, the binary parameters at path are used. + | |||
| `sequence` + | `"transformations":List[Transformation]` + | A sequence of transformations, Applying the sequence applies the composition of all transforms in the list, in order. + | |||
| `displacements` + | `"path":str` `"interpolation":str` + | Displacement field transformation located at (path). + | |||
| `coordinates` + | `"path":str` `"interpolation":str` + | Coordinate field transformation located at (path). + | |||
| `inverseOf` + | `"transform":Transform` + | The inverse of a transformation. Useful if a transform is not closed-form invertible. See Forward and inverse for details and examples. + | |||
| `bijection` + | `"forward":Transform` `"inverse":Transform` + | Explicitly define an invertible transformation by providing a forward transformation and its inverse. + | |||
| `byDimension` + | `"transformations":List[Transformation]` + | Define a high dimensional transformation using lower dimensional transformations on subsets of
+ dimensions.
| type | fields | description
| |
+store.zarr # Root folder of the zarr store +│ +├── .zattrs # coordinate transformations describing the relationship between two image coordinate systems +│ # are stored in the attributes of their parent group. +│ # transformations between 'volume' and 'crop' coordinate systems are stored here. +│ +├── coordinateTransformations # transformations that use array storage go in a "coordinateTransformations" zarr group. +│ └── displacements # for example, a zarr array containing a displacement field +│ ├── .zattrs +│ └── .zarray +│ +├── volume +│ ├── .zattrs # group level attributes (multiscales) +│ └── 0 # a group containing the 0th scale +│ └── image # a zarr array +│ ├── .zattrs # physical coordinate system and transformations here +│ └── .zarray # the array attributes +└── crop + ├── .zattrs # group level attributes (multiscales) + └── 0 # a group containing the 0th scale + └── image # a zarr array + ├── .zattrs # physical coordinate system and transformations here + └── .zarray # the array attributes ++ +### Additional details + +Most coordinate transformations MUST specify their input and output coordinate systems using `input` and `output` with a string value +corresponding to the name of a coordinate system. The coordinate system's name may be the path to an array, and therefore may +not appear in the list of coordinate systems. + +Exceptions are if the the coordinate transformation appears in the `transformations` list of a `sequence` or is the +`transformation` of an `inverseOf` transformation. In these two cases input and output SHOULD be omitted (see below for +details). + +Transformations in the `transformations` list of a `byDimensions` transformation MUST provide `input` and `output` as arrays +of strings corresponding to axis names of the parent transformation's input and output coordinate systems (see below for +details). + +
+path: examples/transformations/identity.json +highlight: json ++ +defines the function: + +``` +x = i +y = j +``` + +
+path: examples/transformations/mapAxis1.json +highlight: json ++ +The "equivalent to identity" transformation defines the function: + +``` +x = i +y = j +``` + +and the "permutation" transformation defines the function + +``` +x = j +y = i +``` + +
+path: examples/transformations/mapAxis2.json +highlight: json ++ +The "projection_down" transformation defines the function: + +``` +x = b +``` + +and the "projection_up" transformation defines the function: + +``` +x = a +y = b +z = b +``` +
+path: examples/transformations/translation.json +highlight: json ++ +defines the function: + +``` +x = i + 9 +y = j - 1.42 +``` +
+path: examples/transformations/scale.json +highlight: json ++ +defines the function: + +``` +x = 3.12 * i +y = 2 * j +``` +
+ path: examples/transformations/affine2d2d.json + highlight: json ++ + defines the function: + + ``` + x = 1*i + 2*j + 3 + y = 4*i + 5*j + 6 + ``` + + it is equivalent to this matrix-vector multiplication in homogeneous coordinates: + + ``` + [ 1 2 3 ][ i ] [ x ] + [ 4 5 6 ][ j ] = [ y ] + [ 0 0 1 ][ 1 ] [ 1 ] + ``` + + where the last row `[0 0 1]` is omitted in the JSON representation. + +
+ path: examples/transformations/affine2d3d.json + highlight: json ++ + defines the function: + + ``` + x = 1*i + 2*j + 3 + y = 4*i + 5*j + 6 + z = 7*i + 8*j + 9 + ``` + + it is equivalent to this matrix-vector multiplication in homogeneous coordinates: + + ``` + [ 1 2 3 ][ i ] [ x ] + [ 4 5 6 ][ j ] = [ y ] + [ 7 8 9 ][ 1 ] [ z ] + [ 0 0 1 ] [ 1 ] + ``` + + where the last row `[0 0 1]` is omitted in the JSON representation. +
+ path: examples/transformations/rotation.json + highlight: json ++ + defines the function: + + ``` + x = 0*i - 1*j + y = 1*i + 0*j + ``` +
+ path: examples/transformations/inverseOf.json + highlight: json ++ +
+path: examples/transformations/sequence.json +highlight: json ++ +describes the function + +``` +x = (i + 0.1) * 2 +y = (j + 0.9) * 3 +``` + +and is invertible. +
linear (default)nearestcubic+path: examples/transformations/byDimension1.json +highlight: json ++ +
+path: examples/transformations/byDimension2.json +highlight: json ++ +
+path: examples/transformations/byDimensionInvalid1.json +highlight: json ++ +It is invalid for two reasons. First because input `0` used by the scale transformation is not an axis of the `byDimension` transformation's `input`. Second, the `x` axis of the `output` does not appear in the `output` of any child transformation. + +
+path: examples/transformations/byDimensionInvalid2.json +highlight: json ++ +This transformation is invalid because the output axis `x` appears in more than one transformation in the `transformations` list. + +
+path: examples/transformations/bijection.json +highlight: json ++ +the input and output of the `forward` and `inverse` transformations are understood to be: + +
+path: examples/transformations/bijection_verbose.json +highlight: json ++ +
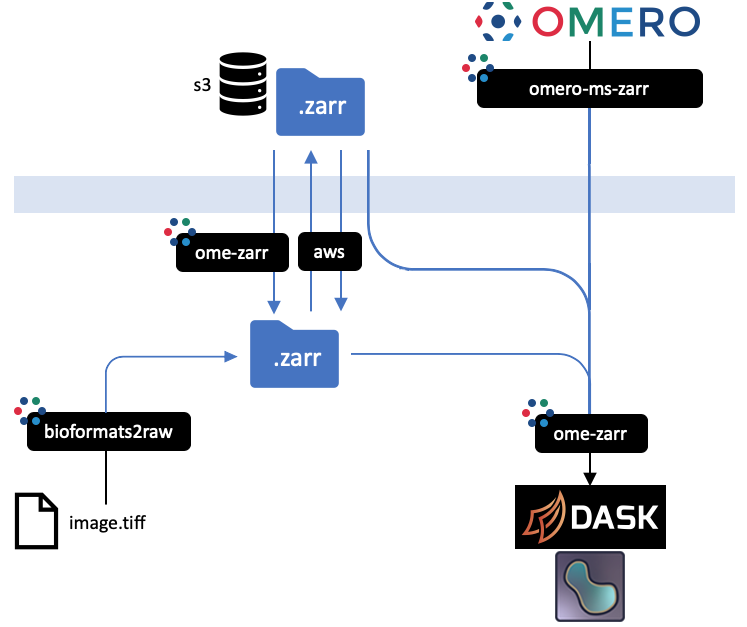 +
+ All implementations prevent an equivalent representation of a dataset which can be downloaded or uploaded freely. An interactive
version of this diagram is available from the [OME2020 Workshop](https://downloads.openmicroscopy.org/presentations/2020/Dundee/Workshops/NGFF/zarr_diagram/).
@@ -785,6 +1832,20 @@ Version History {#history}
"et al"
],
"date": "06 October 2020"
+ },
+ "itk":{
+ "id": "itk-book",
+ "href": "https://itk.org/ItkSoftwareGuide.pdf",
+ "title": "The ITK Software Guide",
+ "status": "Informational",
+ "publisher": "ITK",
+ "authors": [
+ "Hans J. Johnson",
+ "Matthew M. McCormick",
+ "Luis Ibanez",
+ "Insight Software Consortium"
+ ],
+ "date": "16 April 2021"
}
}
diff --git a/latest/schemas/axes.schema b/latest/schemas/axes.schema
new file mode 100644
index 00000000..56b447f2
--- /dev/null
+++ b/latest/schemas/axes.schema
@@ -0,0 +1,62 @@
+{
+ "$schema": "https://json-schema.org/draft/2020-12/schema",
+ "$id": "https://ngff.openmicroscopy.org/latest/schemas/axes.schema",
+ "title": "NGFF Axes",
+ "description": "JSON from OME-NGFF .zattrs",
+ "type": "array",
+ "uniqueItems": true,
+ "minItems": 1,
+ "items": {
+ "$ref": "#/$defs/axis"
+ },
+ "$comment": "Ensure that there exist at most three space axes",
+ "contains": {
+ "type": "object",
+ "properties": {
+ "name": {
+ "type": "string"
+ },
+ "type": {
+ "type": "string",
+ "enum": [
+ "space"
+ ]
+ },
+ "unit": {
+ "type": "string"
+ }
+ }
+ },
+ "minContains": 0,
+ "maxContains": 3,
+ "$defs": {
+ "axis": {
+ "type": "object",
+ "properties": {
+ "name": {
+ "type": "string",
+ "description": "Name of the axis"
+ },
+ "longName": {
+ "type": "string",
+ "description": "Longer name or description of the axis."
+ },
+ "type": {
+ "type": "string",
+ "description": "Dimension of the axis"
+ },
+ "discrete": {
+ "type": "boolean",
+ "description": "Whether the dimension is discrete"
+ },
+ "unit": {
+ "type": "string",
+ "description": "Unit for the axis"
+ }
+ },
+ "required": [
+ "name"
+ ]
+ }
+ }
+}
diff --git a/latest/schemas/coordinateTransformations.schema b/latest/schemas/coordinateTransformations.schema
new file mode 100644
index 00000000..8bd35f4f
--- /dev/null
+++ b/latest/schemas/coordinateTransformations.schema
@@ -0,0 +1,296 @@
+{
+ "$schema": "https://json-schema.org/draft/2020-12/schema",
+ "$id": "https://ngff.openmicroscopy.org/latest/schemas/coordinateTransformations.schema",
+ "title": "NGFF Coordinate Transformations",
+ "description": "JSON from OME-NGFF .zattrs",
+ "type": "array",
+ "minItems": 1,
+ "contains": {
+ "type": "object"
+ },
+ "maxContains": 1,
+ "items": {
+ "$ref": "#/$defs/transformation"
+ },
+ "$defs": {
+ "transformation" : {
+ "oneOf": [
+ {
+ "$ref": "#/$defs/scaleTransformation"
+ },
+ {
+ "$ref": "#/$defs/translationTransformation"
+ },
+ {
+ "$ref": "#/$defs/affineTransformation"
+ },
+ {
+ "$ref": "#/$defs/rotationTransformation"
+ },
+ {
+ "$ref": "#/$defs/inverseOfTransformation"
+ },
+ {
+ "$ref": "#/$defs/sequenceTransformation"
+ },
+ {
+ "$ref": "#/$defs/coordinatesTransformation"
+ },
+ {
+ "$ref": "#/$defs/displacementsTransformation"
+ },
+ {
+ "$ref": "#/$defs/byDimensionTransformation"
+ },
+ {
+ "$ref": "#/$defs/bijectionTransformation"
+ }
+ ]
+ },
+ "scaleTransformation": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "type": "string",
+ "enum": [
+ "scale"
+ ]
+ },
+ "scale": {
+ "type": "array",
+ "minItems": 2,
+ "items": {
+ "type": "number"
+ }
+ },
+ "path": {
+ "type": "string"
+ }
+ },
+ "required": [
+ "type"
+ ],
+ "oneOf": [
+ {
+ "required": ["scale"]
+ },
+ {
+ "required": ["path"]
+ }
+ ]
+ },
+ "translationTransformation": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "type": "string",
+ "enum": [
+ "translation"
+ ]
+ },
+ "translation": {
+ "type": "array",
+ "minItems": 2,
+ "items": {
+ "type": "number"
+ }
+ },
+ "path": {
+ "type": "string"
+ }
+ },
+ "required": [
+ "type"
+ ],
+ "oneOf": [
+ {
+ "required": ["translation"]
+ },
+ {
+ "required": ["path"]
+ }
+ ]
+ },
+ "affineTransformation": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "type": "string",
+ "enum": [
+ "affine"
+ ]
+ },
+ "affine": {
+ "type": "array",
+ "minItems": 2,
+ "items": {
+ "type": "number"
+ }
+ },
+ "path": {
+ "type": "string"
+ }
+ },
+ "required": [
+ "type"
+ ],
+ "oneOf": [
+ {
+ "required": ["affine"]
+ },
+ {
+ "required": ["path"]
+ }
+ ]
+ },
+ "rotationTransformation": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "type": "string",
+ "enum": [
+ "rotation"
+ ]
+ },
+ "rotation": {
+ "type": "object",
+ "minItems": 2,
+ "items": {
+ "type": "number"
+ }
+ },
+ "path": {
+ "type": "string"
+ }
+ },
+ "required": [
+ "type"
+ ],
+ "oneOf": [
+ {
+ "required": ["rotation"]
+ },
+ {
+ "required": ["path"]
+ }
+ ]
+ },
+ "inverseOfTransformation": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "type": "string",
+ "enum": [
+ "inverseOf"
+ ]
+ },
+ "transformation": {
+ "type": "object"
+ }
+ },
+ "required": [
+ "type", "transformation"
+ ]
+ },
+ "sequenceTransformation": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "type": "string",
+ "enum": [
+ "sequence"
+ ]
+ },
+ "transformations": {
+ "type": "array"
+ }
+ },
+ "required": [
+ "type", "transformations"
+ ]
+ },
+ "coordinatesTransformation": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "type": "string",
+ "enum": [
+ "coordinates"
+ ]
+ },
+ "path": {
+ "type": "string"
+ },
+ "interpolation": {
+ "type": "string",
+ "enum": [
+ "nearest", "linear", "cubic"
+ ]
+ }
+ },
+ "required": [
+ "type", "path", "interpolation"
+ ]
+ },
+ "displacementsTransformation": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "type": "string",
+ "enum": [
+ "displacements"
+ ]
+ },
+ "path": {
+ "type": "string"
+ },
+ "interpolation": {
+ "type": "string",
+ "enum": [
+ "nearest", "linear", "cubic"
+ ]
+ }
+ },
+ "required": [
+ "type", "path", "interpolation"
+ ]
+ },
+ "byDimensionTransformation": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "type": "string",
+ "enum": [
+ "byDimension"
+ ]
+ },
+ "transformations": {
+ "type": "array"
+ }
+ },
+ "required": [
+ "type", "transformations"
+ ]
+ },
+ "bijectionTransformation": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "type": "string",
+ "enum": [
+ "bijection"
+ ]
+ },
+ "forward": {
+ "type": "object"
+ },
+ "inverse": {
+ "type": "object"
+ }
+ },
+ "required": [
+ "type", "forward", "inverse"
+ ]
+ }
+ }
+}
diff --git a/latest/schemas/coordinate_systems.schema b/latest/schemas/coordinate_systems.schema
new file mode 100644
index 00000000..583d9941
--- /dev/null
+++ b/latest/schemas/coordinate_systems.schema
@@ -0,0 +1,30 @@
+{
+ "$schema": "https://json-schema.org/draft/2020-12/schema",
+ "$id": "https://ngff.openmicroscopy.org/latest/schemas/coordinate_systems.schema",
+ "title": "NGFF CoordinateSystem",
+ "description": "JSON from OME-NGFF .zattrs",
+ "type": "array",
+ "uniqueItems": true,
+ "items": {
+ "$ref": "#/$defs/coordinateSystem"
+ },
+ "$defs": {
+ "coordinateSystem": {
+ "description": "Coordinate Systems for OME-NGFF",
+ "type": "object",
+ "properties": {
+ "name": {
+ "type": "string",
+ "description": "Name of coordinate system"
+ },
+ "axes": {
+ "$ref": "axes.schema"
+ }
+ },
+ "required": [
+ "name",
+ "axes"
+ ]
+ }
+ }
+}
diff --git a/latest/schemas/coordinate_systems_and_transforms.schema b/latest/schemas/coordinate_systems_and_transforms.schema
new file mode 100644
index 00000000..403f78a5
--- /dev/null
+++ b/latest/schemas/coordinate_systems_and_transforms.schema
@@ -0,0 +1,45 @@
+{
+ "$schema": "https://json-schema.org/draft/2020-12/schema",
+ "$id": "https://ngff.openmicroscopy.org/latest/schemas/coordinate_systems_and_transforms.schema",
+ "title": "NGFF Coordinate Systems and Transforms",
+ "description": "Coordinate Systems and transforms for OME-NGFF",
+ "type": "object",
+ "properties": {
+ "coordinateSystems": {
+ "$ref": "coordinate_systems.schema"
+ },
+ "coordinateTransformations": {
+ "$ref": "coordinate_transformation.schema"
+ },
+ "arrayCoordinateSystem": {
+ "type": "object",
+ "properties": {
+ "name": {
+ "type": "string",
+ "description": "Name of coordinate space"
+ },
+ "axes": {
+ "allOf": [
+ {
+ "$ref": "axes.schema"
+ },
+ {
+ "type": "array",
+ "items": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "const": "array"
+ }
+ }
+ }
+ }
+ ]
+ }
+ },
+ "required": [
+ "axes"
+ ]
+ }
+ }
+}
diff --git a/latest/schemas/coordinate_transformation.schema b/latest/schemas/coordinate_transformation.schema
new file mode 100644
index 00000000..e5156da0
--- /dev/null
+++ b/latest/schemas/coordinate_transformation.schema
@@ -0,0 +1,344 @@
+{
+ "$schema": "https://json-schema.org/draft/2020-12/schema",
+ "$id": "https://ngff.openmicroscopy.org/latest/schemas/coordinate_transformation.schema",
+ "title": "NGFF Coordinate Systems and Transforms",
+ "description": "Coordinate Systems and transforms for OME-NGFF",
+ "type": "array",
+ "uniqueItems": true,
+ "minItems": 1,
+ "items": {
+ "allOf": [
+ {
+ "$ref": "#/$defs/coordinateTransformation"
+ },
+ {
+ "type": "object",
+ "properties": {
+ "input": {
+ "type": "string"
+ },
+ "output": {
+ "type": "string"
+ }
+ },
+ "required": [
+ "input",
+ "output"
+ ]
+ }
+ ]
+ },
+ "$defs": {
+ "path_w_url": {
+ "description": "Path specification. Schema local solution until https://github.com/ome/ngff/issues/144 is resolved.",
+ "type": "object",
+ "properties": {
+ "path": {
+ "type": "string"
+ },
+ "url": {
+ "type": "string",
+ "format": "uri"
+ }
+ },
+ "required": [
+ "path"
+ ]
+ },
+ "coordinateTransformation": {
+ "description": "OME-NGFF coordinate transformation.",
+ "allOf": [
+ {
+ "type": "object",
+ "properties": {
+ "name": {
+ "type": "string"
+ },
+ "type": {
+ "type": "string"
+ }
+ },
+ "required": [
+ "type"
+ ]
+ },
+ {
+ "oneOf": [
+ {
+ "$ref": "#/$defs/identity"
+ },
+ {
+ "$ref": "#/$defs/mapAxis"
+ },
+ {
+ "$ref": "#/$defs/scale"
+ },
+ {
+ "$ref": "#/$defs/translation"
+ },
+ {
+ "$ref": "#/$defs/affine"
+ },
+ {
+ "$ref": "#/$defs/rotation"
+ },
+ {
+ "$ref": "#/$defs/inverseOf"
+ },
+ {
+ "$ref": "#/$defs/bijection"
+ },
+ {
+ "$ref": "#/$defs/sequence"
+ },
+ {
+ "$ref": "#/$defs/byDimension"
+ },
+ {
+ "$ref": "#/$defs/displacements"
+ },
+ {
+ "$ref": "#/$defs/coordinates"
+ }
+ ]
+ }
+ ]
+ },
+ "byDimensionTransformation": {
+ "type": "object",
+ "description": "Transformation used inside a byDimension transformation",
+ "allOf": [
+ { "$ref": "#/$defs/coordinateTransformation" },
+ {
+ "properties": {
+ "input": {
+ "type": "array",
+ "items": {
+ "type": "string"
+ }
+ },
+ "output": {
+ "type": "array",
+ "items": {
+ "type": "string"
+ }
+ }
+ }
+ }
+ ],
+ "required": ["input", "output"]
+ },
+ "identity": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "const": "identity"
+ }
+ }
+ },
+ "mapAxis": {
+ "type": "object",
+ "description": "Permute axes by name",
+ "properties": {
+ "type": {
+ "const": "mapAxis"
+ },
+ "mapAxis": {
+ "type": "object",
+ "patternProperties": {
+ ".*": {
+ "type": "string"
+ }
+ }
+ },
+ "required": [
+ "mapAxis"
+ ]
+ }
+ },
+ "scale": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "const": "scale"
+ }
+ },
+ "oneOf": [
+ {
+ "$ref": "#/$defs/path_w_url"
+ },
+ {
+ "properties": {
+ "scale": {
+ "type": "array",
+ "items": {
+ "type": "number",
+ "exclusiveMinimum": 0
+ }
+ }
+ },
+ "required": [
+ "scale"
+ ]
+ }
+ ]
+ },
+ "translation": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "const": "translation"
+ }
+ },
+ "oneOf": [
+ {
+ "$ref": "#/$defs/path_w_url"
+ },
+ {
+ "properties": {
+ "translation": {
+ "type": "array",
+ "items": {
+ "type": "number"
+ }
+ }
+ },
+ "required": [
+ "translation"
+ ]
+ }
+ ]
+ },
+ "mtxFlatOrNested" : {
+ "type": "array",
+ "oneOf": [
+ {
+ "items": { "type": "number" }
+ },
+ {
+ "items": {
+ "type": "array",
+ "items" : { "type" : "number" }
+ }
+ }
+ ]
+ },
+ "affine": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "const": "affine"
+ }
+ },
+ "oneOf": [
+ {
+ "$ref": "#/$defs/path_w_url"
+ },
+ {
+ "properties": {
+ "affine": {
+ "$ref": "#/$defs/mtxFlatOrNested"
+ },
+ "required": [
+ "affine"
+ ]
+ }
+ }
+ ]
+ },
+ "rotation": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "const": "rotation"
+ }
+ },
+ "oneOf": [
+ {
+ "$ref": "#/$defs/path_w_url"
+ },
+ {
+ "properties": {
+ "rotation": {
+ "type": "array",
+ "items": {
+ "type": "number"
+ }
+ },
+ "required": [
+ "rotation"
+ ]
+ }
+ }
+ ]
+ },
+ "inverseOf": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "const": "inverseOf"
+ },
+ "transformation": {
+ "$ref": "#/$defs/coordinateTransformation"
+ }
+ },
+ "required": [
+ "transformation"
+ ]
+ },
+ "bijection": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "const": "bijection"
+ },
+ "forward": {
+ "$ref": "#/$defs/coordinateTransformation"
+ },
+ "inverse": {
+ "$ref": "#/$defs/coordinateTransformation"
+ }
+ },
+ "required": [
+ "forward", "inverse"
+ ]
+ },
+ "sequence": {
+ "description": "A sequence of transformations",
+ "type": "object",
+ "properties": {
+ "type": { "const": "sequence" },
+ "transformations": {
+ "type": "array",
+ "items": {
+ "$ref": "#/$defs/coordinateTransformation"
+ }
+ }
+ }
+ },
+ "byDimension": {
+ "type": "object",
+ "properties": {
+ "type": { "const": "byDimension" },
+ "transformations": {
+ "type": "array",
+ "items": {
+ "$ref": "#/$defs/byDimensionTransformation"
+ }
+ }
+ }
+ },
+ "displacements": {
+ "type": "object",
+ "properties": {
+ "type": { "const": "displacements" }
+ }
+ },
+ "coordinates": {
+ "type": "object",
+ "properties": {
+ "type": { "const": "coordinates" }
+ }
+ }
+ }
+}
diff --git a/latest/schemas/image.schema b/latest/schemas/image.schema
index 3926c4f4..4dd3954e 100644
--- a/latest/schemas/image.schema
+++ b/latest/schemas/image.schema
@@ -24,7 +24,7 @@
"type": "string"
},
"coordinateTransformations": {
- "$ref": "#/$defs/coordinateTransformations"
+ "$ref": "coordinate_transformation.schema"
}
},
"required": ["path", "coordinateTransformations"]
@@ -36,15 +36,15 @@
"0.5-dev"
]
},
- "axes": {
- "$ref": "#/$defs/axes"
+ "coordinateSystems": {
+ "$ref": "coordinate_systems.schema"
},
"coordinateTransformations": {
- "$ref": "#/$defs/coordinateTransformations"
- }
+ "$ref": "coordinate_transformation.schema"
+ }
},
"required": [
- "datasets", "axes"
+ "datasets", "coordinateSystems"
]
},
"minItems": 1,
@@ -106,128 +106,5 @@
]
}
},
- "required": [ "multiscales" ],
-
- "$defs": {
- "axes": {
- "type": "array",
- "uniqueItems": true,
- "minItems": 2,
- "maxItems": 5,
- "contains": {
- "type": "object",
- "properties": {
- "name": {
- "type": "string"
- },
- "type": {
- "type": "string",
- "enum": ["space"]
- },
- "units": {
- "type": "string"
- }
- }
- },
- "minContains": 2,
- "maxContains": 3,
- "items": {
- "oneOf": [
- {
- "type": "object",
- "properties": {
- "name": {
- "type": "string"
- },
- "type": {
- "type": "string",
- "enum": ["channel", "time", "space"]
- }
- },
- "required": ["name", "type"]
- },
- {
- "type": "object",
- "properties": {
- "name": {
- "type": "string"
- },
- "type": {
- "type": "string",
- "not": {
- "enum": ["space", "time", "channel"]
- }
- }
- },
- "required": ["name"]
- }
- ]
- }
- },
- "coordinateTransformations": {
- "type": "array",
- "minItems": 1,
- "contains": {
- "type": "object",
- "properties": {
- "type": {
- "type": "string",
- "enum": [
- "scale"
- ]
- },
- "scale": {
- "type": "array",
- "minItems": 2,
- "items": {
- "type": "number"
- }
- }
- }
- },
- "maxContains": 1,
- "items": {
- "oneOf": [
- {
- "type": "object",
- "properties": {
- "type": {
- "type": "string",
- "enum": [
- "scale"
- ]
- },
- "scale": {
- "type": "array",
- "minItems": 2,
- "items": {
- "type": "number"
- }
- }
- },
- "required": ["type", "scale"]
- },
- {
- "type": "object",
- "properties": {
- "type": {
- "type": "string",
- "enum": [
- "translation"
- ]
- },
- "translation": {
- "type": "array",
- "minItems": 2,
- "items": {
- "type": "number"
- }
- }
- },
- "required": ["type", "translation"]
- }
- ]
- }
- }
- }
+ "required": [ "multiscales" ]
}
diff --git a/latest/schemas/strict_axes.schema b/latest/schemas/strict_axes.schema
new file mode 100644
index 00000000..522602d2
--- /dev/null
+++ b/latest/schemas/strict_axes.schema
@@ -0,0 +1,30 @@
+{
+ "$schema": "https://json-schema.org/draft/2020-12/schema",
+ "$id": "https://ngff.openmicroscopy.org/latest/schemas/strict_axes.schema",
+ "title": "NGFF Strict Axes",
+ "description": "JSON from OME-NGFF .zattrs",
+ "allOf": [
+ {
+ "$ref": "https://ngff.openmicroscopy.org/latest/schemas/axes.schema"
+ },
+ {
+ "items": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "type": "string",
+ "enum": [
+ "array",
+ "channel",
+ "time",
+ "space",
+ "displacement",
+ "coordinate",
+ "frequency"
+ ]
+ }
+ }
+ }
+ }
+ ]
+}
diff --git a/latest/schemas/strict_coordinate_systems.schema b/latest/schemas/strict_coordinate_systems.schema
new file mode 100644
index 00000000..425ccbe5
--- /dev/null
+++ b/latest/schemas/strict_coordinate_systems.schema
@@ -0,0 +1,18 @@
+{
+ "$id": "https://ngff.openmicroscopy.org/latest/schemas/strict_coordinate_systems.schema",
+ "allOf" : [
+ {
+ "$ref": "coordinate_systems.schema"
+ },
+ {
+ "items": {
+ "type": "object",
+ "properties": {
+ "axes": {
+ "$ref": "strict_axes.schema"
+ }
+ }
+ }
+ }
+ ]
+}
diff --git a/latest/schemas/strict_image.schema b/latest/schemas/strict_image.schema
index bcecc003..4f631ba2 100644
--- a/latest/schemas/strict_image.schema
+++ b/latest/schemas/strict_image.schema
@@ -8,6 +8,11 @@
"properties": {
"multiscales": {
"items": {
+ "properties" : {
+ "coordinateSystems": {
+ "$ref": "strict_coordinate_systems.schema"
+ }
+ },
"required": [
"version", "metadata", "type", "name"
]
@@ -16,4 +21,4 @@
}
}
]
-}
\ No newline at end of file
+}
diff --git a/latest/tests/image_suite.json b/latest/tests/image_suite.json
index 39cb73d5..e017dac0 100644
--- a/latest/tests/image_suite.json
+++ b/latest/tests/image_suite.json
@@ -10,23 +10,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "t",
- "type": "time",
- "units": "micrometer"
- },
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
@@ -36,12 +19,36 @@
0.13,
0.13
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "mismatch_axes_units"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "mismatch_axes_units",
+ "axes": [
+ {
+ "name": "t",
+ "type": "time",
+ "unit": "micrometer"
+ },
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
@@ -53,21 +60,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "angle"
- },
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
@@ -78,12 +70,34 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "untyped_axes"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "untyped_axes",
+ "axes": [
+ {
+ "name": "angle"
+ },
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
@@ -106,21 +120,28 @@
"scale": [
1,
1
- ]
+ ],
+ "input": "/0",
+ "output": "missing_version"
}
]
}
],
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
+ "coordinateSystems": [
{
- "name": "x",
- "type": "space",
- "units": "micrometer"
+ "name": "missing_version",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
}
]
}
@@ -134,18 +155,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micron"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
@@ -155,12 +164,31 @@
0.13,
0.13
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "invalid_axis_units"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "invalid_axis_units",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micron"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
@@ -183,7 +211,9 @@
"scale": [
1,
1
- ]
+ ],
+ "input": "/0",
+ "output": "invalid_axis_units"
}
]
}
@@ -200,16 +230,21 @@
"multichannel": true
}
},
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
+ "coordinateSystems": [
{
- "name": "x",
- "type": "space",
- "units": "micrometer"
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
}
]
}
@@ -223,22 +258,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "angle",
- "type": "custom"
- },
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
@@ -249,12 +268,35 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "custom_type_axes"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "custom_type_axes",
+ "axes": [
+ {
+ "name": "angle",
+ "type": "custom"
+ },
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
@@ -266,18 +308,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
@@ -287,33 +317,42 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "custom_type_axes"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "duplicate_axes",
+ "axes": [
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
"valid": false
},
{
- "formerly": "invalid/missing_space_axes.json",
+ "formerly": "valid/no_space_axes.json",
"description": "TBD",
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "t",
- "type": "time"
- },
- {
- "name": "c",
- "type": "channel"
- }
- ],
"datasets": [
{
"path": "0",
@@ -323,16 +362,33 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "no_space_axes"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "no_space_axes",
+ "axes": [
+ {
+ "name": "t",
+ "type": "time"
+ },
+ {
+ "name": "c",
+ "type": "channel"
+ }
+ ]
+ }
+ ]
}
]
},
- "valid": false
+ "valid": true
},
{
"formerly": "invalid/invalid_transformation_type.json",
@@ -340,18 +396,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
@@ -361,35 +405,42 @@
1,
1
],
- "type": "translation"
+ "type": "translation",
+ "input": "/0",
+ "output": "transformation_type"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "transformation_type",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
"valid": false
},
{
- "formerly": "invalid/missing_scale.json",
+ "formerly": "invalid/translation_only.json",
"description": "TBD",
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
@@ -399,49 +450,42 @@
1,
1
],
- "type": "translation"
+ "type": "translation",
+ "input": "/0",
+ "output": "transformation_type"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "transformation_type",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
- "valid": false
+ "valid": true
},
{
- "formerly": "invalid/too_many_axes.json",
+ "formerly": "valid/many_axes.json",
"description": "TBD",
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "angle",
- "type": "custom"
- },
- {
- "name": "t",
- "type": "time"
- },
- {
- "name": "c",
- "type": "channel"
- },
- {
- "name": "z",
- "type": "space"
- },
- {
- "name": "y",
- "type": "space"
- },
- {
- "name": "x",
- "type": "space"
- }
- ],
"datasets": [
{
"path": "0",
@@ -455,16 +499,49 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "many_axes"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "many_axes",
+ "axes": [
+ {
+ "name": "angle",
+ "type": "custom"
+ },
+ {
+ "name": "t",
+ "type": "time"
+ },
+ {
+ "name": "c",
+ "type": "channel"
+ },
+ {
+ "name": "z",
+ "type": "space"
+ },
+ {
+ "name": "y",
+ "type": "space"
+ },
+ {
+ "name": "x",
+ "type": "space"
+ }
+ ]
+ }
+ ]
}
]
},
- "valid": false
+ "valid": true
},
{
"formerly": "invalid/invalid_channels_color.json",
@@ -472,18 +549,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
@@ -493,27 +558,46 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "invalid_channels_color"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "invalie_channels_color",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
],
"omero": {
"channels": [
{
"active": true,
- "coefficient": 1.0,
+ "coefficient": 1,
"color": 255,
"family": "linear",
"label": "1234",
"window": {
- "end": 1765.0,
- "max": 2555.0,
- "min": 5.0,
- "start": 0.0
+ "end": 1765,
+ "max": 2555,
+ "min": 5,
+ "start": 0
}
}
]
@@ -527,16 +611,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "type": "space",
- "units": "micron"
- },
- {
- "type": "space",
- "units": "micron"
- }
- ],
"datasets": [
{
"path": "0",
@@ -546,30 +620,40 @@
0.13,
0.13
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "missing_axes_name"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "missing_axes_name",
+ "axes": [
+ {
+ "type": "space",
+ "unit": "micron"
+ },
+ {
+ "type": "space",
+ "unit": "micron"
+ }
+ ]
+ }
+ ]
}
]
},
"valid": false
},
{
- "formerly": "invalid/invalid_axes_count.json",
+ "formerly": "valid/one_axis.json",
"description": "TBD",
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
@@ -579,37 +663,37 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "one_axis"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "one_axis",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
- "valid": false
+ "valid": true
},
{
- "formerly": "invalid/one_space_axes.json",
+ "formerly": "valid/one_space_axes.json",
"description": "TBD",
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "t",
- "type": "time"
- },
- {
- "name": "c",
- "type": "channel"
- },
- {
- "name": "x",
- "type": "space"
- }
- ],
"datasets": [
{
"path": "0",
@@ -620,16 +704,37 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "out"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "t",
+ "type": "time"
+ },
+ {
+ "name": "c",
+ "type": "channel"
+ },
+ {
+ "name": "x",
+ "type": "space"
+ }
+ ]
+ }
+ ]
}
]
},
- "valid": false
+ "valid": true
},
{
"formerly": "invalid/invalid_path.json",
@@ -637,18 +742,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": 0,
@@ -658,12 +751,31 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "out"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
@@ -675,18 +787,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
@@ -696,7 +796,9 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "out"
}
]
}
@@ -706,10 +808,29 @@
"scale": [
"invalid"
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "out"
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
@@ -721,24 +842,29 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0"
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
@@ -750,20 +876,25 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
+ "datasets": [],
+ "version": "0.5-dev",
+ "coordinateSystems": [
{
- "name": "x",
- "type": "space",
- "units": "micrometer"
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
}
- ],
- "datasets": [],
- "version": "0.5-dev"
+ ]
}
]
},
@@ -775,19 +906,24 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
+ "version": "0.5-dev",
+ "coordinateSystems": [
{
- "name": "x",
- "type": "space",
- "units": "micrometer"
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
}
- ],
- "version": "0.5-dev"
+ ]
}
]
},
@@ -808,7 +944,9 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "out"
}
]
}
@@ -825,18 +963,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
@@ -846,12 +972,31 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "out"
}
]
}
],
- "version": "0.3"
+ "version": "0.3",
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
@@ -863,18 +1008,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "invalid",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
@@ -884,69 +1017,99 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "out"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "y",
+ "type": "invalid",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
- "valid": false
+ "valid": true
},
{
- "formerly": "invalid/duplicate_scale.json",
- "description": "TBD",
+ "formerly": "valid/two_scales.json",
+ "description": "A sequence of two scales. Valid, but not recommended.",
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
"coordinateTransformations": [
{
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
+ "type": "sequence",
+ "input": "/0",
+ "output": "out",
+ "transformations": [
+ {
+ "scale": [
+ 1,
+ 1
+ ],
+ "type": "scale"
+ },
+ {
+ "scale": [
+ 1,
+ 1
+ ],
+ "type": "scale"
+ }
+ ]
+ }
+ ]
+ }
+ ],
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
},
{
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
}
]
}
- ],
- "version": "0.5-dev"
+ ]
}
]
},
- "valid": false
+ "valid": true
},
{
- "formerly": "invalid/no_axes.json",
+ "formerly": "invalid/no_coordinateSystems.json",
"description": "TBD",
"data": {
"multiscales": [
{
- "axes": [],
"datasets": [
{
"path": "0",
@@ -956,12 +1119,20 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "out"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": []
+ }
+ ]
}
]
},
@@ -973,24 +1144,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "X",
- "type": "space"
- },
- {
- "name": "z",
- "type": "space"
- },
- {
- "name": "y",
- "type": "space"
- },
- {
- "name": "x",
- "type": "space"
- }
- ],
"datasets": [
{
"path": "0",
@@ -1002,12 +1155,37 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "out"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "X",
+ "type": "space"
+ },
+ {
+ "name": "z",
+ "type": "space"
+ },
+ {
+ "name": "y",
+ "type": "space"
+ },
+ {
+ "name": "x",
+ "type": "space"
+ }
+ ]
+ }
+ ]
}
]
},
@@ -1028,18 +1206,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
@@ -1049,27 +1215,46 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "out"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
],
"omero": {
"channels": [
{
"active": true,
- "coefficient": 1.0,
+ "coefficient": 1,
"color": "ff0000",
"family": "linear",
"label": "1234",
"window": {
"end": "100",
- "max": 2555.0,
- "min": 5.0,
- "start": 0.0
+ "max": 2555,
+ "min": 5,
+ "start": 0
}
}
]
@@ -1083,25 +1268,30 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
"coordinateTransformations": []
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
@@ -1113,18 +1303,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"coordinateTransformations": [
@@ -1133,12 +1311,31 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "missing_path"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
diff --git a/latest/tests/strict_image_suite.json b/latest/tests/strict_image_suite.json
index b8aeebe1..d55069fb 100644
--- a/latest/tests/strict_image_suite.json
+++ b/latest/tests/strict_image_suite.json
@@ -12,31 +12,65 @@
{
"version": "0.5-dev",
"name": "example",
- "axes": [
- {
- "name": "t",
- "type": "time",
- "unit": "millisecond"
- },
- {
- "name": "c",
- "type": "channel"
- },
- {
- "name": "z",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
+ "coordinateSystems": [
+ {
+ "name": "example",
+ "axes": [
+ {
+ "name": "t",
+ "type": "time",
+ "unit": "millisecond"
+ },
+ {
+ "name": "c",
+ "type": "channel"
+ },
+ {
+ "name": "z",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ },
+ {
+ "name": "tmp",
+ "axes": [
+ {
+ "name": "t",
+ "type": "time",
+ "unit": "millisecond"
+ },
+ {
+ "name": "c",
+ "type": "channel"
+ },
+ {
+ "name": "z",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
],
"datasets": [
{
@@ -45,12 +79,14 @@
{
"type": "scale",
"scale": [
- 1.0,
- 1.0,
+ 1,
+ 1,
0.5,
0.5,
0.5
- ]
+ ],
+ "input": "/0",
+ "output": "tmp"
}
]
},
@@ -65,7 +101,9 @@
1.0,
1.0,
1.0
- ]
+ ],
+ "input": "/1",
+ "output": "tmp"
}
]
},
@@ -80,7 +118,9 @@
2.0,
2.0,
2.0
- ]
+ ],
+ "input": "/2",
+ "output": "tmp"
}
]
}
@@ -94,7 +134,9 @@
1.0,
1.0,
1.0
- ]
+ ],
+ "input": "tmp",
+ "output": "example"
}
],
"type": "gaussian",
@@ -106,30 +148,128 @@
"kwargs": {
"multichannel": true
}
- }
+ },
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "t",
+ "type": "time",
+ "unit": "millisecond"
+ },
+ {
+ "name": "c",
+ "type": "channel"
+ },
+ {
+ "name": "z",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
"valid": true
},
{
- "formerly": "valid_strict/multiscales_transformations.json",
+ "formerly": "invalid_strict/multiscales_nonstandard_axis_type.json",
"description": "TBD",
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
+ "version": "0.5-dev",
+ "name": "example",
+ "coordinateSystems": [
+ {
+ "name": "nonstandard_axis",
+ "axes": [
+ {
+ "name": "c",
+ "type": "custom",
+ "unit": ""
+ },
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ],
+ "datasets": [
{
- "name": "x",
- "type": "space",
- "units": "micrometer"
+ "path": "0",
+ "coordinateTransformations": [
+ {
+ "type": "scale",
+ "scale": [
+ 1.0,
+ 1.0,
+ 1.0
+ ],
+ "input": "/0",
+ "output": "nonstandard_axis"
+ }
+ ]
}
],
+ "type": "gaussian",
+ "metadata": {
+ "description": "the fields in metadata depend on the downscaling implementation. Here, the parameters passed to the skimage function are given",
+ "method": "skimage.transform.pyramid_gaussian",
+ "version": "0.16.1",
+ "args": "[true]",
+ "kwargs": {
+ "multichannel": true
+ }
+ }
+ }
+ ]
+ },
+ "valid": false
+ },
+ {
+ "formerly": "valid_strict/multiscales_transformations.json",
+ "description": "TBD",
+ "data": {
+ "multiscales": [
+ {
+ "coordinateSystems": [
+ {
+ "name": "multiscales_transformations",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ],
"datasets": [
{
"path": "0",
@@ -139,7 +279,9 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "example"
}
]
}
@@ -150,7 +292,9 @@
10,
10
],
- "type": "scale"
+ "type": "scale",
+ "input": "out",
+ "output": "multiscales_transformations"
}
],
"version": "0.5-dev",
@@ -158,7 +302,24 @@
"type": "foo",
"metadata": {
"key": "value"
- }
+ },
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
@@ -184,7 +345,9 @@
"scale": [
1,
1
- ]
+ ],
+ "input": "/path/to/0",
+ "output": "image_metadata"
}
]
}
@@ -201,17 +364,22 @@
"multichannel": true
}
},
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
+ "coordinateSystems": [
+ {
+ "name": "image_metadata",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
]
}
]
@@ -224,17 +392,22 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
+ "coordinateSystems": [
+ {
+ "name": "image",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
],
"datasets": [
{
@@ -245,7 +418,9 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "image"
}
]
}
@@ -255,7 +430,24 @@
"type": "foo",
"metadata": {
"key": "value"
- }
+ },
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
@@ -267,54 +459,66 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "t",
- "type": "time"
- },
- {
- "name": "c",
- "type": "channel"
- },
- {
- "name": "z",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
+ "coordinateSystems": [
+ {
+ "name": "image_omero",
+ "axes": [
+ {
+ "name": "t",
+ "type": "time"
+ },
+ {
+ "name": "c",
+ "type": "channel"
+ },
+ {
+ "name": "z",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
],
"datasets": [
{
"path": "0",
"coordinateTransformations": [
{
- "scale": [
- 1,
- 1,
- 0.5,
- 0.13,
- 0.13
- ],
- "type": "scale"
- },
- {
- "translation": [
- 0,
- 9,
- 0.5,
- 25.74,
- 21.58
- ],
- "type": "translation"
+ "type": "sequence",
+ "transformations" : [
+ {
+ "scale": [
+ 1,
+ 1,
+ 0.5,
+ 0.13,
+ 0.13
+ ],
+ "type": "scale"
+ },
+ {
+ "translation": [
+ 0,
+ 9,
+ 0.5,
+ 25.74,
+ 21.58
+ ],
+ "type": "translation"
+ }
+ ],
+ "input": "/0",
+ "output": "image_omero"
}
]
},
@@ -329,7 +533,9 @@
0.26,
0.26
],
- "type": "scale"
+ "type": "scale",
+ "input": "/1",
+ "output": "image_omero"
}
]
}
@@ -339,37 +545,67 @@
"type": "foo",
"metadata": {
"key": "value"
- }
+ },
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "t",
+ "type": "time"
+ },
+ {
+ "name": "c",
+ "type": "channel"
+ },
+ {
+ "name": "z",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
],
"omero": {
"channels": [
{
"active": true,
- "coefficient": 1.0,
+ "coefficient": 1,
"color": "00FF00",
"family": "linear",
"inverted": false,
"label": "FITC",
"window": {
- "end": 813.0,
- "max": 870.0,
- "min": 102.0,
- "start": 82.0
+ "end": 813,
+ "max": 870,
+ "min": 102,
+ "start": 82
}
},
{
"active": true,
- "coefficient": 1.0,
+ "coefficient": 1,
"color": "FF0000",
"family": "linear",
"inverted": false,
"label": "RD-TR-PE",
"window": {
- "end": 815.0,
- "max": 441.0,
- "min": 129.0,
- "start": 78.0
+ "end": 815,
+ "max": 441,
+ "min": 129,
+ "start": 78
}
}
],
diff --git a/latest/tests/test_validation.py b/latest/tests/test_validation.py
index 8c13b113..d631bd10 100644
--- a/latest/tests/test_validation.py
+++ b/latest/tests/test_validation.py
@@ -49,6 +49,11 @@ def pytest_generate_tests(metafunc):
if "suite" in metafunc.fixturenames:
suites: List[Schema] = []
ids: List[str] = []
+ schema_store = {}
+ for filename in glob.glob("schemas/*.schema"):
+ with open(filename) as o:
+ schema = json.load(o)
+ schema_store[schema["$id"]] = schema
# Validation
for filename in glob.glob("tests/*.json"):
diff --git a/latest/transform-details.bs b/latest/transform-details.bs
new file mode 100644
index 00000000..b2b744c8
--- /dev/null
+++ b/latest/transform-details.bs
@@ -0,0 +1,106 @@
+
All implementations prevent an equivalent representation of a dataset which can be downloaded or uploaded freely. An interactive
version of this diagram is available from the [OME2020 Workshop](https://downloads.openmicroscopy.org/presentations/2020/Dundee/Workshops/NGFF/zarr_diagram/).
@@ -785,6 +1832,20 @@ Version History {#history}
"et al"
],
"date": "06 October 2020"
+ },
+ "itk":{
+ "id": "itk-book",
+ "href": "https://itk.org/ItkSoftwareGuide.pdf",
+ "title": "The ITK Software Guide",
+ "status": "Informational",
+ "publisher": "ITK",
+ "authors": [
+ "Hans J. Johnson",
+ "Matthew M. McCormick",
+ "Luis Ibanez",
+ "Insight Software Consortium"
+ ],
+ "date": "16 April 2021"
}
}
diff --git a/latest/schemas/axes.schema b/latest/schemas/axes.schema
new file mode 100644
index 00000000..56b447f2
--- /dev/null
+++ b/latest/schemas/axes.schema
@@ -0,0 +1,62 @@
+{
+ "$schema": "https://json-schema.org/draft/2020-12/schema",
+ "$id": "https://ngff.openmicroscopy.org/latest/schemas/axes.schema",
+ "title": "NGFF Axes",
+ "description": "JSON from OME-NGFF .zattrs",
+ "type": "array",
+ "uniqueItems": true,
+ "minItems": 1,
+ "items": {
+ "$ref": "#/$defs/axis"
+ },
+ "$comment": "Ensure that there exist at most three space axes",
+ "contains": {
+ "type": "object",
+ "properties": {
+ "name": {
+ "type": "string"
+ },
+ "type": {
+ "type": "string",
+ "enum": [
+ "space"
+ ]
+ },
+ "unit": {
+ "type": "string"
+ }
+ }
+ },
+ "minContains": 0,
+ "maxContains": 3,
+ "$defs": {
+ "axis": {
+ "type": "object",
+ "properties": {
+ "name": {
+ "type": "string",
+ "description": "Name of the axis"
+ },
+ "longName": {
+ "type": "string",
+ "description": "Longer name or description of the axis."
+ },
+ "type": {
+ "type": "string",
+ "description": "Dimension of the axis"
+ },
+ "discrete": {
+ "type": "boolean",
+ "description": "Whether the dimension is discrete"
+ },
+ "unit": {
+ "type": "string",
+ "description": "Unit for the axis"
+ }
+ },
+ "required": [
+ "name"
+ ]
+ }
+ }
+}
diff --git a/latest/schemas/coordinateTransformations.schema b/latest/schemas/coordinateTransformations.schema
new file mode 100644
index 00000000..8bd35f4f
--- /dev/null
+++ b/latest/schemas/coordinateTransformations.schema
@@ -0,0 +1,296 @@
+{
+ "$schema": "https://json-schema.org/draft/2020-12/schema",
+ "$id": "https://ngff.openmicroscopy.org/latest/schemas/coordinateTransformations.schema",
+ "title": "NGFF Coordinate Transformations",
+ "description": "JSON from OME-NGFF .zattrs",
+ "type": "array",
+ "minItems": 1,
+ "contains": {
+ "type": "object"
+ },
+ "maxContains": 1,
+ "items": {
+ "$ref": "#/$defs/transformation"
+ },
+ "$defs": {
+ "transformation" : {
+ "oneOf": [
+ {
+ "$ref": "#/$defs/scaleTransformation"
+ },
+ {
+ "$ref": "#/$defs/translationTransformation"
+ },
+ {
+ "$ref": "#/$defs/affineTransformation"
+ },
+ {
+ "$ref": "#/$defs/rotationTransformation"
+ },
+ {
+ "$ref": "#/$defs/inverseOfTransformation"
+ },
+ {
+ "$ref": "#/$defs/sequenceTransformation"
+ },
+ {
+ "$ref": "#/$defs/coordinatesTransformation"
+ },
+ {
+ "$ref": "#/$defs/displacementsTransformation"
+ },
+ {
+ "$ref": "#/$defs/byDimensionTransformation"
+ },
+ {
+ "$ref": "#/$defs/bijectionTransformation"
+ }
+ ]
+ },
+ "scaleTransformation": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "type": "string",
+ "enum": [
+ "scale"
+ ]
+ },
+ "scale": {
+ "type": "array",
+ "minItems": 2,
+ "items": {
+ "type": "number"
+ }
+ },
+ "path": {
+ "type": "string"
+ }
+ },
+ "required": [
+ "type"
+ ],
+ "oneOf": [
+ {
+ "required": ["scale"]
+ },
+ {
+ "required": ["path"]
+ }
+ ]
+ },
+ "translationTransformation": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "type": "string",
+ "enum": [
+ "translation"
+ ]
+ },
+ "translation": {
+ "type": "array",
+ "minItems": 2,
+ "items": {
+ "type": "number"
+ }
+ },
+ "path": {
+ "type": "string"
+ }
+ },
+ "required": [
+ "type"
+ ],
+ "oneOf": [
+ {
+ "required": ["translation"]
+ },
+ {
+ "required": ["path"]
+ }
+ ]
+ },
+ "affineTransformation": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "type": "string",
+ "enum": [
+ "affine"
+ ]
+ },
+ "affine": {
+ "type": "array",
+ "minItems": 2,
+ "items": {
+ "type": "number"
+ }
+ },
+ "path": {
+ "type": "string"
+ }
+ },
+ "required": [
+ "type"
+ ],
+ "oneOf": [
+ {
+ "required": ["affine"]
+ },
+ {
+ "required": ["path"]
+ }
+ ]
+ },
+ "rotationTransformation": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "type": "string",
+ "enum": [
+ "rotation"
+ ]
+ },
+ "rotation": {
+ "type": "object",
+ "minItems": 2,
+ "items": {
+ "type": "number"
+ }
+ },
+ "path": {
+ "type": "string"
+ }
+ },
+ "required": [
+ "type"
+ ],
+ "oneOf": [
+ {
+ "required": ["rotation"]
+ },
+ {
+ "required": ["path"]
+ }
+ ]
+ },
+ "inverseOfTransformation": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "type": "string",
+ "enum": [
+ "inverseOf"
+ ]
+ },
+ "transformation": {
+ "type": "object"
+ }
+ },
+ "required": [
+ "type", "transformation"
+ ]
+ },
+ "sequenceTransformation": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "type": "string",
+ "enum": [
+ "sequence"
+ ]
+ },
+ "transformations": {
+ "type": "array"
+ }
+ },
+ "required": [
+ "type", "transformations"
+ ]
+ },
+ "coordinatesTransformation": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "type": "string",
+ "enum": [
+ "coordinates"
+ ]
+ },
+ "path": {
+ "type": "string"
+ },
+ "interpolation": {
+ "type": "string",
+ "enum": [
+ "nearest", "linear", "cubic"
+ ]
+ }
+ },
+ "required": [
+ "type", "path", "interpolation"
+ ]
+ },
+ "displacementsTransformation": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "type": "string",
+ "enum": [
+ "displacements"
+ ]
+ },
+ "path": {
+ "type": "string"
+ },
+ "interpolation": {
+ "type": "string",
+ "enum": [
+ "nearest", "linear", "cubic"
+ ]
+ }
+ },
+ "required": [
+ "type", "path", "interpolation"
+ ]
+ },
+ "byDimensionTransformation": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "type": "string",
+ "enum": [
+ "byDimension"
+ ]
+ },
+ "transformations": {
+ "type": "array"
+ }
+ },
+ "required": [
+ "type", "transformations"
+ ]
+ },
+ "bijectionTransformation": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "type": "string",
+ "enum": [
+ "bijection"
+ ]
+ },
+ "forward": {
+ "type": "object"
+ },
+ "inverse": {
+ "type": "object"
+ }
+ },
+ "required": [
+ "type", "forward", "inverse"
+ ]
+ }
+ }
+}
diff --git a/latest/schemas/coordinate_systems.schema b/latest/schemas/coordinate_systems.schema
new file mode 100644
index 00000000..583d9941
--- /dev/null
+++ b/latest/schemas/coordinate_systems.schema
@@ -0,0 +1,30 @@
+{
+ "$schema": "https://json-schema.org/draft/2020-12/schema",
+ "$id": "https://ngff.openmicroscopy.org/latest/schemas/coordinate_systems.schema",
+ "title": "NGFF CoordinateSystem",
+ "description": "JSON from OME-NGFF .zattrs",
+ "type": "array",
+ "uniqueItems": true,
+ "items": {
+ "$ref": "#/$defs/coordinateSystem"
+ },
+ "$defs": {
+ "coordinateSystem": {
+ "description": "Coordinate Systems for OME-NGFF",
+ "type": "object",
+ "properties": {
+ "name": {
+ "type": "string",
+ "description": "Name of coordinate system"
+ },
+ "axes": {
+ "$ref": "axes.schema"
+ }
+ },
+ "required": [
+ "name",
+ "axes"
+ ]
+ }
+ }
+}
diff --git a/latest/schemas/coordinate_systems_and_transforms.schema b/latest/schemas/coordinate_systems_and_transforms.schema
new file mode 100644
index 00000000..403f78a5
--- /dev/null
+++ b/latest/schemas/coordinate_systems_and_transforms.schema
@@ -0,0 +1,45 @@
+{
+ "$schema": "https://json-schema.org/draft/2020-12/schema",
+ "$id": "https://ngff.openmicroscopy.org/latest/schemas/coordinate_systems_and_transforms.schema",
+ "title": "NGFF Coordinate Systems and Transforms",
+ "description": "Coordinate Systems and transforms for OME-NGFF",
+ "type": "object",
+ "properties": {
+ "coordinateSystems": {
+ "$ref": "coordinate_systems.schema"
+ },
+ "coordinateTransformations": {
+ "$ref": "coordinate_transformation.schema"
+ },
+ "arrayCoordinateSystem": {
+ "type": "object",
+ "properties": {
+ "name": {
+ "type": "string",
+ "description": "Name of coordinate space"
+ },
+ "axes": {
+ "allOf": [
+ {
+ "$ref": "axes.schema"
+ },
+ {
+ "type": "array",
+ "items": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "const": "array"
+ }
+ }
+ }
+ }
+ ]
+ }
+ },
+ "required": [
+ "axes"
+ ]
+ }
+ }
+}
diff --git a/latest/schemas/coordinate_transformation.schema b/latest/schemas/coordinate_transformation.schema
new file mode 100644
index 00000000..e5156da0
--- /dev/null
+++ b/latest/schemas/coordinate_transformation.schema
@@ -0,0 +1,344 @@
+{
+ "$schema": "https://json-schema.org/draft/2020-12/schema",
+ "$id": "https://ngff.openmicroscopy.org/latest/schemas/coordinate_transformation.schema",
+ "title": "NGFF Coordinate Systems and Transforms",
+ "description": "Coordinate Systems and transforms for OME-NGFF",
+ "type": "array",
+ "uniqueItems": true,
+ "minItems": 1,
+ "items": {
+ "allOf": [
+ {
+ "$ref": "#/$defs/coordinateTransformation"
+ },
+ {
+ "type": "object",
+ "properties": {
+ "input": {
+ "type": "string"
+ },
+ "output": {
+ "type": "string"
+ }
+ },
+ "required": [
+ "input",
+ "output"
+ ]
+ }
+ ]
+ },
+ "$defs": {
+ "path_w_url": {
+ "description": "Path specification. Schema local solution until https://github.com/ome/ngff/issues/144 is resolved.",
+ "type": "object",
+ "properties": {
+ "path": {
+ "type": "string"
+ },
+ "url": {
+ "type": "string",
+ "format": "uri"
+ }
+ },
+ "required": [
+ "path"
+ ]
+ },
+ "coordinateTransformation": {
+ "description": "OME-NGFF coordinate transformation.",
+ "allOf": [
+ {
+ "type": "object",
+ "properties": {
+ "name": {
+ "type": "string"
+ },
+ "type": {
+ "type": "string"
+ }
+ },
+ "required": [
+ "type"
+ ]
+ },
+ {
+ "oneOf": [
+ {
+ "$ref": "#/$defs/identity"
+ },
+ {
+ "$ref": "#/$defs/mapAxis"
+ },
+ {
+ "$ref": "#/$defs/scale"
+ },
+ {
+ "$ref": "#/$defs/translation"
+ },
+ {
+ "$ref": "#/$defs/affine"
+ },
+ {
+ "$ref": "#/$defs/rotation"
+ },
+ {
+ "$ref": "#/$defs/inverseOf"
+ },
+ {
+ "$ref": "#/$defs/bijection"
+ },
+ {
+ "$ref": "#/$defs/sequence"
+ },
+ {
+ "$ref": "#/$defs/byDimension"
+ },
+ {
+ "$ref": "#/$defs/displacements"
+ },
+ {
+ "$ref": "#/$defs/coordinates"
+ }
+ ]
+ }
+ ]
+ },
+ "byDimensionTransformation": {
+ "type": "object",
+ "description": "Transformation used inside a byDimension transformation",
+ "allOf": [
+ { "$ref": "#/$defs/coordinateTransformation" },
+ {
+ "properties": {
+ "input": {
+ "type": "array",
+ "items": {
+ "type": "string"
+ }
+ },
+ "output": {
+ "type": "array",
+ "items": {
+ "type": "string"
+ }
+ }
+ }
+ }
+ ],
+ "required": ["input", "output"]
+ },
+ "identity": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "const": "identity"
+ }
+ }
+ },
+ "mapAxis": {
+ "type": "object",
+ "description": "Permute axes by name",
+ "properties": {
+ "type": {
+ "const": "mapAxis"
+ },
+ "mapAxis": {
+ "type": "object",
+ "patternProperties": {
+ ".*": {
+ "type": "string"
+ }
+ }
+ },
+ "required": [
+ "mapAxis"
+ ]
+ }
+ },
+ "scale": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "const": "scale"
+ }
+ },
+ "oneOf": [
+ {
+ "$ref": "#/$defs/path_w_url"
+ },
+ {
+ "properties": {
+ "scale": {
+ "type": "array",
+ "items": {
+ "type": "number",
+ "exclusiveMinimum": 0
+ }
+ }
+ },
+ "required": [
+ "scale"
+ ]
+ }
+ ]
+ },
+ "translation": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "const": "translation"
+ }
+ },
+ "oneOf": [
+ {
+ "$ref": "#/$defs/path_w_url"
+ },
+ {
+ "properties": {
+ "translation": {
+ "type": "array",
+ "items": {
+ "type": "number"
+ }
+ }
+ },
+ "required": [
+ "translation"
+ ]
+ }
+ ]
+ },
+ "mtxFlatOrNested" : {
+ "type": "array",
+ "oneOf": [
+ {
+ "items": { "type": "number" }
+ },
+ {
+ "items": {
+ "type": "array",
+ "items" : { "type" : "number" }
+ }
+ }
+ ]
+ },
+ "affine": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "const": "affine"
+ }
+ },
+ "oneOf": [
+ {
+ "$ref": "#/$defs/path_w_url"
+ },
+ {
+ "properties": {
+ "affine": {
+ "$ref": "#/$defs/mtxFlatOrNested"
+ },
+ "required": [
+ "affine"
+ ]
+ }
+ }
+ ]
+ },
+ "rotation": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "const": "rotation"
+ }
+ },
+ "oneOf": [
+ {
+ "$ref": "#/$defs/path_w_url"
+ },
+ {
+ "properties": {
+ "rotation": {
+ "type": "array",
+ "items": {
+ "type": "number"
+ }
+ },
+ "required": [
+ "rotation"
+ ]
+ }
+ }
+ ]
+ },
+ "inverseOf": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "const": "inverseOf"
+ },
+ "transformation": {
+ "$ref": "#/$defs/coordinateTransformation"
+ }
+ },
+ "required": [
+ "transformation"
+ ]
+ },
+ "bijection": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "const": "bijection"
+ },
+ "forward": {
+ "$ref": "#/$defs/coordinateTransformation"
+ },
+ "inverse": {
+ "$ref": "#/$defs/coordinateTransformation"
+ }
+ },
+ "required": [
+ "forward", "inverse"
+ ]
+ },
+ "sequence": {
+ "description": "A sequence of transformations",
+ "type": "object",
+ "properties": {
+ "type": { "const": "sequence" },
+ "transformations": {
+ "type": "array",
+ "items": {
+ "$ref": "#/$defs/coordinateTransformation"
+ }
+ }
+ }
+ },
+ "byDimension": {
+ "type": "object",
+ "properties": {
+ "type": { "const": "byDimension" },
+ "transformations": {
+ "type": "array",
+ "items": {
+ "$ref": "#/$defs/byDimensionTransformation"
+ }
+ }
+ }
+ },
+ "displacements": {
+ "type": "object",
+ "properties": {
+ "type": { "const": "displacements" }
+ }
+ },
+ "coordinates": {
+ "type": "object",
+ "properties": {
+ "type": { "const": "coordinates" }
+ }
+ }
+ }
+}
diff --git a/latest/schemas/image.schema b/latest/schemas/image.schema
index 3926c4f4..4dd3954e 100644
--- a/latest/schemas/image.schema
+++ b/latest/schemas/image.schema
@@ -24,7 +24,7 @@
"type": "string"
},
"coordinateTransformations": {
- "$ref": "#/$defs/coordinateTransformations"
+ "$ref": "coordinate_transformation.schema"
}
},
"required": ["path", "coordinateTransformations"]
@@ -36,15 +36,15 @@
"0.5-dev"
]
},
- "axes": {
- "$ref": "#/$defs/axes"
+ "coordinateSystems": {
+ "$ref": "coordinate_systems.schema"
},
"coordinateTransformations": {
- "$ref": "#/$defs/coordinateTransformations"
- }
+ "$ref": "coordinate_transformation.schema"
+ }
},
"required": [
- "datasets", "axes"
+ "datasets", "coordinateSystems"
]
},
"minItems": 1,
@@ -106,128 +106,5 @@
]
}
},
- "required": [ "multiscales" ],
-
- "$defs": {
- "axes": {
- "type": "array",
- "uniqueItems": true,
- "minItems": 2,
- "maxItems": 5,
- "contains": {
- "type": "object",
- "properties": {
- "name": {
- "type": "string"
- },
- "type": {
- "type": "string",
- "enum": ["space"]
- },
- "units": {
- "type": "string"
- }
- }
- },
- "minContains": 2,
- "maxContains": 3,
- "items": {
- "oneOf": [
- {
- "type": "object",
- "properties": {
- "name": {
- "type": "string"
- },
- "type": {
- "type": "string",
- "enum": ["channel", "time", "space"]
- }
- },
- "required": ["name", "type"]
- },
- {
- "type": "object",
- "properties": {
- "name": {
- "type": "string"
- },
- "type": {
- "type": "string",
- "not": {
- "enum": ["space", "time", "channel"]
- }
- }
- },
- "required": ["name"]
- }
- ]
- }
- },
- "coordinateTransformations": {
- "type": "array",
- "minItems": 1,
- "contains": {
- "type": "object",
- "properties": {
- "type": {
- "type": "string",
- "enum": [
- "scale"
- ]
- },
- "scale": {
- "type": "array",
- "minItems": 2,
- "items": {
- "type": "number"
- }
- }
- }
- },
- "maxContains": 1,
- "items": {
- "oneOf": [
- {
- "type": "object",
- "properties": {
- "type": {
- "type": "string",
- "enum": [
- "scale"
- ]
- },
- "scale": {
- "type": "array",
- "minItems": 2,
- "items": {
- "type": "number"
- }
- }
- },
- "required": ["type", "scale"]
- },
- {
- "type": "object",
- "properties": {
- "type": {
- "type": "string",
- "enum": [
- "translation"
- ]
- },
- "translation": {
- "type": "array",
- "minItems": 2,
- "items": {
- "type": "number"
- }
- }
- },
- "required": ["type", "translation"]
- }
- ]
- }
- }
- }
+ "required": [ "multiscales" ]
}
diff --git a/latest/schemas/strict_axes.schema b/latest/schemas/strict_axes.schema
new file mode 100644
index 00000000..522602d2
--- /dev/null
+++ b/latest/schemas/strict_axes.schema
@@ -0,0 +1,30 @@
+{
+ "$schema": "https://json-schema.org/draft/2020-12/schema",
+ "$id": "https://ngff.openmicroscopy.org/latest/schemas/strict_axes.schema",
+ "title": "NGFF Strict Axes",
+ "description": "JSON from OME-NGFF .zattrs",
+ "allOf": [
+ {
+ "$ref": "https://ngff.openmicroscopy.org/latest/schemas/axes.schema"
+ },
+ {
+ "items": {
+ "type": "object",
+ "properties": {
+ "type": {
+ "type": "string",
+ "enum": [
+ "array",
+ "channel",
+ "time",
+ "space",
+ "displacement",
+ "coordinate",
+ "frequency"
+ ]
+ }
+ }
+ }
+ }
+ ]
+}
diff --git a/latest/schemas/strict_coordinate_systems.schema b/latest/schemas/strict_coordinate_systems.schema
new file mode 100644
index 00000000..425ccbe5
--- /dev/null
+++ b/latest/schemas/strict_coordinate_systems.schema
@@ -0,0 +1,18 @@
+{
+ "$id": "https://ngff.openmicroscopy.org/latest/schemas/strict_coordinate_systems.schema",
+ "allOf" : [
+ {
+ "$ref": "coordinate_systems.schema"
+ },
+ {
+ "items": {
+ "type": "object",
+ "properties": {
+ "axes": {
+ "$ref": "strict_axes.schema"
+ }
+ }
+ }
+ }
+ ]
+}
diff --git a/latest/schemas/strict_image.schema b/latest/schemas/strict_image.schema
index bcecc003..4f631ba2 100644
--- a/latest/schemas/strict_image.schema
+++ b/latest/schemas/strict_image.schema
@@ -8,6 +8,11 @@
"properties": {
"multiscales": {
"items": {
+ "properties" : {
+ "coordinateSystems": {
+ "$ref": "strict_coordinate_systems.schema"
+ }
+ },
"required": [
"version", "metadata", "type", "name"
]
@@ -16,4 +21,4 @@
}
}
]
-}
\ No newline at end of file
+}
diff --git a/latest/tests/image_suite.json b/latest/tests/image_suite.json
index 39cb73d5..e017dac0 100644
--- a/latest/tests/image_suite.json
+++ b/latest/tests/image_suite.json
@@ -10,23 +10,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "t",
- "type": "time",
- "units": "micrometer"
- },
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
@@ -36,12 +19,36 @@
0.13,
0.13
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "mismatch_axes_units"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "mismatch_axes_units",
+ "axes": [
+ {
+ "name": "t",
+ "type": "time",
+ "unit": "micrometer"
+ },
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
@@ -53,21 +60,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "angle"
- },
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
@@ -78,12 +70,34 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "untyped_axes"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "untyped_axes",
+ "axes": [
+ {
+ "name": "angle"
+ },
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
@@ -106,21 +120,28 @@
"scale": [
1,
1
- ]
+ ],
+ "input": "/0",
+ "output": "missing_version"
}
]
}
],
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
+ "coordinateSystems": [
{
- "name": "x",
- "type": "space",
- "units": "micrometer"
+ "name": "missing_version",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
}
]
}
@@ -134,18 +155,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micron"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
@@ -155,12 +164,31 @@
0.13,
0.13
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "invalid_axis_units"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "invalid_axis_units",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micron"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
@@ -183,7 +211,9 @@
"scale": [
1,
1
- ]
+ ],
+ "input": "/0",
+ "output": "invalid_axis_units"
}
]
}
@@ -200,16 +230,21 @@
"multichannel": true
}
},
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
+ "coordinateSystems": [
{
- "name": "x",
- "type": "space",
- "units": "micrometer"
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
}
]
}
@@ -223,22 +258,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "angle",
- "type": "custom"
- },
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
@@ -249,12 +268,35 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "custom_type_axes"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "custom_type_axes",
+ "axes": [
+ {
+ "name": "angle",
+ "type": "custom"
+ },
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
@@ -266,18 +308,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
@@ -287,33 +317,42 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "custom_type_axes"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "duplicate_axes",
+ "axes": [
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
"valid": false
},
{
- "formerly": "invalid/missing_space_axes.json",
+ "formerly": "valid/no_space_axes.json",
"description": "TBD",
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "t",
- "type": "time"
- },
- {
- "name": "c",
- "type": "channel"
- }
- ],
"datasets": [
{
"path": "0",
@@ -323,16 +362,33 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "no_space_axes"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "no_space_axes",
+ "axes": [
+ {
+ "name": "t",
+ "type": "time"
+ },
+ {
+ "name": "c",
+ "type": "channel"
+ }
+ ]
+ }
+ ]
}
]
},
- "valid": false
+ "valid": true
},
{
"formerly": "invalid/invalid_transformation_type.json",
@@ -340,18 +396,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
@@ -361,35 +405,42 @@
1,
1
],
- "type": "translation"
+ "type": "translation",
+ "input": "/0",
+ "output": "transformation_type"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "transformation_type",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
"valid": false
},
{
- "formerly": "invalid/missing_scale.json",
+ "formerly": "invalid/translation_only.json",
"description": "TBD",
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
@@ -399,49 +450,42 @@
1,
1
],
- "type": "translation"
+ "type": "translation",
+ "input": "/0",
+ "output": "transformation_type"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "transformation_type",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
- "valid": false
+ "valid": true
},
{
- "formerly": "invalid/too_many_axes.json",
+ "formerly": "valid/many_axes.json",
"description": "TBD",
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "angle",
- "type": "custom"
- },
- {
- "name": "t",
- "type": "time"
- },
- {
- "name": "c",
- "type": "channel"
- },
- {
- "name": "z",
- "type": "space"
- },
- {
- "name": "y",
- "type": "space"
- },
- {
- "name": "x",
- "type": "space"
- }
- ],
"datasets": [
{
"path": "0",
@@ -455,16 +499,49 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "many_axes"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "many_axes",
+ "axes": [
+ {
+ "name": "angle",
+ "type": "custom"
+ },
+ {
+ "name": "t",
+ "type": "time"
+ },
+ {
+ "name": "c",
+ "type": "channel"
+ },
+ {
+ "name": "z",
+ "type": "space"
+ },
+ {
+ "name": "y",
+ "type": "space"
+ },
+ {
+ "name": "x",
+ "type": "space"
+ }
+ ]
+ }
+ ]
}
]
},
- "valid": false
+ "valid": true
},
{
"formerly": "invalid/invalid_channels_color.json",
@@ -472,18 +549,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
@@ -493,27 +558,46 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "invalid_channels_color"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "invalie_channels_color",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
],
"omero": {
"channels": [
{
"active": true,
- "coefficient": 1.0,
+ "coefficient": 1,
"color": 255,
"family": "linear",
"label": "1234",
"window": {
- "end": 1765.0,
- "max": 2555.0,
- "min": 5.0,
- "start": 0.0
+ "end": 1765,
+ "max": 2555,
+ "min": 5,
+ "start": 0
}
}
]
@@ -527,16 +611,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "type": "space",
- "units": "micron"
- },
- {
- "type": "space",
- "units": "micron"
- }
- ],
"datasets": [
{
"path": "0",
@@ -546,30 +620,40 @@
0.13,
0.13
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "missing_axes_name"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "missing_axes_name",
+ "axes": [
+ {
+ "type": "space",
+ "unit": "micron"
+ },
+ {
+ "type": "space",
+ "unit": "micron"
+ }
+ ]
+ }
+ ]
}
]
},
"valid": false
},
{
- "formerly": "invalid/invalid_axes_count.json",
+ "formerly": "valid/one_axis.json",
"description": "TBD",
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
@@ -579,37 +663,37 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "one_axis"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "one_axis",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
- "valid": false
+ "valid": true
},
{
- "formerly": "invalid/one_space_axes.json",
+ "formerly": "valid/one_space_axes.json",
"description": "TBD",
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "t",
- "type": "time"
- },
- {
- "name": "c",
- "type": "channel"
- },
- {
- "name": "x",
- "type": "space"
- }
- ],
"datasets": [
{
"path": "0",
@@ -620,16 +704,37 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "out"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "t",
+ "type": "time"
+ },
+ {
+ "name": "c",
+ "type": "channel"
+ },
+ {
+ "name": "x",
+ "type": "space"
+ }
+ ]
+ }
+ ]
}
]
},
- "valid": false
+ "valid": true
},
{
"formerly": "invalid/invalid_path.json",
@@ -637,18 +742,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": 0,
@@ -658,12 +751,31 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "out"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
@@ -675,18 +787,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
@@ -696,7 +796,9 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "out"
}
]
}
@@ -706,10 +808,29 @@
"scale": [
"invalid"
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "out"
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
@@ -721,24 +842,29 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0"
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
@@ -750,20 +876,25 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
+ "datasets": [],
+ "version": "0.5-dev",
+ "coordinateSystems": [
{
- "name": "x",
- "type": "space",
- "units": "micrometer"
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
}
- ],
- "datasets": [],
- "version": "0.5-dev"
+ ]
}
]
},
@@ -775,19 +906,24 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
+ "version": "0.5-dev",
+ "coordinateSystems": [
{
- "name": "x",
- "type": "space",
- "units": "micrometer"
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
}
- ],
- "version": "0.5-dev"
+ ]
}
]
},
@@ -808,7 +944,9 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "out"
}
]
}
@@ -825,18 +963,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
@@ -846,12 +972,31 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "out"
}
]
}
],
- "version": "0.3"
+ "version": "0.3",
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
@@ -863,18 +1008,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "invalid",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
@@ -884,69 +1017,99 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "out"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "y",
+ "type": "invalid",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
- "valid": false
+ "valid": true
},
{
- "formerly": "invalid/duplicate_scale.json",
- "description": "TBD",
+ "formerly": "valid/two_scales.json",
+ "description": "A sequence of two scales. Valid, but not recommended.",
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
"coordinateTransformations": [
{
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
+ "type": "sequence",
+ "input": "/0",
+ "output": "out",
+ "transformations": [
+ {
+ "scale": [
+ 1,
+ 1
+ ],
+ "type": "scale"
+ },
+ {
+ "scale": [
+ 1,
+ 1
+ ],
+ "type": "scale"
+ }
+ ]
+ }
+ ]
+ }
+ ],
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
},
{
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
}
]
}
- ],
- "version": "0.5-dev"
+ ]
}
]
},
- "valid": false
+ "valid": true
},
{
- "formerly": "invalid/no_axes.json",
+ "formerly": "invalid/no_coordinateSystems.json",
"description": "TBD",
"data": {
"multiscales": [
{
- "axes": [],
"datasets": [
{
"path": "0",
@@ -956,12 +1119,20 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "out"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": []
+ }
+ ]
}
]
},
@@ -973,24 +1144,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "X",
- "type": "space"
- },
- {
- "name": "z",
- "type": "space"
- },
- {
- "name": "y",
- "type": "space"
- },
- {
- "name": "x",
- "type": "space"
- }
- ],
"datasets": [
{
"path": "0",
@@ -1002,12 +1155,37 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "out"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "X",
+ "type": "space"
+ },
+ {
+ "name": "z",
+ "type": "space"
+ },
+ {
+ "name": "y",
+ "type": "space"
+ },
+ {
+ "name": "x",
+ "type": "space"
+ }
+ ]
+ }
+ ]
}
]
},
@@ -1028,18 +1206,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
@@ -1049,27 +1215,46 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "out"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
],
"omero": {
"channels": [
{
"active": true,
- "coefficient": 1.0,
+ "coefficient": 1,
"color": "ff0000",
"family": "linear",
"label": "1234",
"window": {
"end": "100",
- "max": 2555.0,
- "min": 5.0,
- "start": 0.0
+ "max": 2555,
+ "min": 5,
+ "start": 0
}
}
]
@@ -1083,25 +1268,30 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"path": "0",
"coordinateTransformations": []
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
@@ -1113,18 +1303,6 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
- ],
"datasets": [
{
"coordinateTransformations": [
@@ -1133,12 +1311,31 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "missing_path"
}
]
}
],
- "version": "0.5-dev"
+ "version": "0.5-dev",
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
diff --git a/latest/tests/strict_image_suite.json b/latest/tests/strict_image_suite.json
index b8aeebe1..d55069fb 100644
--- a/latest/tests/strict_image_suite.json
+++ b/latest/tests/strict_image_suite.json
@@ -12,31 +12,65 @@
{
"version": "0.5-dev",
"name": "example",
- "axes": [
- {
- "name": "t",
- "type": "time",
- "unit": "millisecond"
- },
- {
- "name": "c",
- "type": "channel"
- },
- {
- "name": "z",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
+ "coordinateSystems": [
+ {
+ "name": "example",
+ "axes": [
+ {
+ "name": "t",
+ "type": "time",
+ "unit": "millisecond"
+ },
+ {
+ "name": "c",
+ "type": "channel"
+ },
+ {
+ "name": "z",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ },
+ {
+ "name": "tmp",
+ "axes": [
+ {
+ "name": "t",
+ "type": "time",
+ "unit": "millisecond"
+ },
+ {
+ "name": "c",
+ "type": "channel"
+ },
+ {
+ "name": "z",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
],
"datasets": [
{
@@ -45,12 +79,14 @@
{
"type": "scale",
"scale": [
- 1.0,
- 1.0,
+ 1,
+ 1,
0.5,
0.5,
0.5
- ]
+ ],
+ "input": "/0",
+ "output": "tmp"
}
]
},
@@ -65,7 +101,9 @@
1.0,
1.0,
1.0
- ]
+ ],
+ "input": "/1",
+ "output": "tmp"
}
]
},
@@ -80,7 +118,9 @@
2.0,
2.0,
2.0
- ]
+ ],
+ "input": "/2",
+ "output": "tmp"
}
]
}
@@ -94,7 +134,9 @@
1.0,
1.0,
1.0
- ]
+ ],
+ "input": "tmp",
+ "output": "example"
}
],
"type": "gaussian",
@@ -106,30 +148,128 @@
"kwargs": {
"multichannel": true
}
- }
+ },
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "t",
+ "type": "time",
+ "unit": "millisecond"
+ },
+ {
+ "name": "c",
+ "type": "channel"
+ },
+ {
+ "name": "z",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
"valid": true
},
{
- "formerly": "valid_strict/multiscales_transformations.json",
+ "formerly": "invalid_strict/multiscales_nonstandard_axis_type.json",
"description": "TBD",
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
+ "version": "0.5-dev",
+ "name": "example",
+ "coordinateSystems": [
+ {
+ "name": "nonstandard_axis",
+ "axes": [
+ {
+ "name": "c",
+ "type": "custom",
+ "unit": ""
+ },
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ],
+ "datasets": [
{
- "name": "x",
- "type": "space",
- "units": "micrometer"
+ "path": "0",
+ "coordinateTransformations": [
+ {
+ "type": "scale",
+ "scale": [
+ 1.0,
+ 1.0,
+ 1.0
+ ],
+ "input": "/0",
+ "output": "nonstandard_axis"
+ }
+ ]
}
],
+ "type": "gaussian",
+ "metadata": {
+ "description": "the fields in metadata depend on the downscaling implementation. Here, the parameters passed to the skimage function are given",
+ "method": "skimage.transform.pyramid_gaussian",
+ "version": "0.16.1",
+ "args": "[true]",
+ "kwargs": {
+ "multichannel": true
+ }
+ }
+ }
+ ]
+ },
+ "valid": false
+ },
+ {
+ "formerly": "valid_strict/multiscales_transformations.json",
+ "description": "TBD",
+ "data": {
+ "multiscales": [
+ {
+ "coordinateSystems": [
+ {
+ "name": "multiscales_transformations",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ],
"datasets": [
{
"path": "0",
@@ -139,7 +279,9 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "example"
}
]
}
@@ -150,7 +292,9 @@
10,
10
],
- "type": "scale"
+ "type": "scale",
+ "input": "out",
+ "output": "multiscales_transformations"
}
],
"version": "0.5-dev",
@@ -158,7 +302,24 @@
"type": "foo",
"metadata": {
"key": "value"
- }
+ },
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
@@ -184,7 +345,9 @@
"scale": [
1,
1
- ]
+ ],
+ "input": "/path/to/0",
+ "output": "image_metadata"
}
]
}
@@ -201,17 +364,22 @@
"multichannel": true
}
},
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
+ "coordinateSystems": [
+ {
+ "name": "image_metadata",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
]
}
]
@@ -224,17 +392,22 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
+ "coordinateSystems": [
+ {
+ "name": "image",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
],
"datasets": [
{
@@ -245,7 +418,9 @@
1,
1
],
- "type": "scale"
+ "type": "scale",
+ "input": "/0",
+ "output": "image"
}
]
}
@@ -255,7 +430,24 @@
"type": "foo",
"metadata": {
"key": "value"
- }
+ },
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
]
},
@@ -267,54 +459,66 @@
"data": {
"multiscales": [
{
- "axes": [
- {
- "name": "t",
- "type": "time"
- },
- {
- "name": "c",
- "type": "channel"
- },
- {
- "name": "z",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "y",
- "type": "space",
- "units": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "units": "micrometer"
- }
+ "coordinateSystems": [
+ {
+ "name": "image_omero",
+ "axes": [
+ {
+ "name": "t",
+ "type": "time"
+ },
+ {
+ "name": "c",
+ "type": "channel"
+ },
+ {
+ "name": "z",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
],
"datasets": [
{
"path": "0",
"coordinateTransformations": [
{
- "scale": [
- 1,
- 1,
- 0.5,
- 0.13,
- 0.13
- ],
- "type": "scale"
- },
- {
- "translation": [
- 0,
- 9,
- 0.5,
- 25.74,
- 21.58
- ],
- "type": "translation"
+ "type": "sequence",
+ "transformations" : [
+ {
+ "scale": [
+ 1,
+ 1,
+ 0.5,
+ 0.13,
+ 0.13
+ ],
+ "type": "scale"
+ },
+ {
+ "translation": [
+ 0,
+ 9,
+ 0.5,
+ 25.74,
+ 21.58
+ ],
+ "type": "translation"
+ }
+ ],
+ "input": "/0",
+ "output": "image_omero"
}
]
},
@@ -329,7 +533,9 @@
0.26,
0.26
],
- "type": "scale"
+ "type": "scale",
+ "input": "/1",
+ "output": "image_omero"
}
]
}
@@ -339,37 +545,67 @@
"type": "foo",
"metadata": {
"key": "value"
- }
+ },
+ "coordinateSystems": [
+ {
+ "name": "tmpname",
+ "axes": [
+ {
+ "name": "t",
+ "type": "time"
+ },
+ {
+ "name": "c",
+ "type": "channel"
+ },
+ {
+ "name": "z",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
}
],
"omero": {
"channels": [
{
"active": true,
- "coefficient": 1.0,
+ "coefficient": 1,
"color": "00FF00",
"family": "linear",
"inverted": false,
"label": "FITC",
"window": {
- "end": 813.0,
- "max": 870.0,
- "min": 102.0,
- "start": 82.0
+ "end": 813,
+ "max": 870,
+ "min": 102,
+ "start": 82
}
},
{
"active": true,
- "coefficient": 1.0,
+ "coefficient": 1,
"color": "FF0000",
"family": "linear",
"inverted": false,
"label": "RD-TR-PE",
"window": {
- "end": 815.0,
- "max": 441.0,
- "min": 129.0,
- "start": 78.0
+ "end": 815,
+ "max": 441,
+ "min": 129,
+ "start": 78
}
}
],
diff --git a/latest/tests/test_validation.py b/latest/tests/test_validation.py
index 8c13b113..d631bd10 100644
--- a/latest/tests/test_validation.py
+++ b/latest/tests/test_validation.py
@@ -49,6 +49,11 @@ def pytest_generate_tests(metafunc):
if "suite" in metafunc.fixturenames:
suites: List[Schema] = []
ids: List[str] = []
+ schema_store = {}
+ for filename in glob.glob("schemas/*.schema"):
+ with open(filename) as o:
+ schema = json.load(o)
+ schema_store[schema["$id"]] = schema
# Validation
for filename in glob.glob("tests/*.json"):
diff --git a/latest/transform-details.bs b/latest/transform-details.bs
new file mode 100644
index 00000000..b2b744c8
--- /dev/null
+++ b/latest/transform-details.bs
@@ -0,0 +1,106 @@
++Title: Coordinates and Transformations +Shortname: ome-ngff-transformations +Level: 1 +Status: LS-COMMIT +Status: w3c/ED +Group: ome +URL: https://ngff.openmicroscopy.org/latest/ +Repository: https://github.com/ome/ngff +Issue Tracking: Forums https://forum.image.sc/tag/ome-ngff +Logo: http://www.openmicroscopy.org/img/logos/ome-logomark.svg +Local Boilerplate: header no +Local Boilerplate: copyright no +Boilerplate: style-darkmode off +Markup Shorthands: markdown yes +Editor: Josh Moore, Open Microscopy Environment (OME) https://www.openmicroscopy.org +Editor: Sébastien Besson, Open Microscopy Environment (OME) https://www.openmicroscopy.org +Editor: Constantin Pape, European Molecular Biology Laboratory (EMBL) https://www.embl.org/sites/heidelberg/ +Editor: John Bogovic, Hughes Medical Institute Janelia (HHMI) https://www.janelia.org/ +Abstract: This document contains next-generation file format (NGFF) +Abstract: specifications for storing bioimaging data in the cloud. +Abstract: All specifications are submitted to the https://image.sc community for review. +Status Text: The current released version of this specification is +Status Text: will be provided between numbered versions. Data written with these latest changes +Status Text: (an "editor's draft") will not necessarily be supported. ++ +Coordinates and Axes {#coords-axes} +===================== + +OME-NGFF datasets are arrays that hold values. The arrays may be indexed by discrete (integer) +coordinates in order to obtain a corresponding value. If values are desired at continuous (real-valued) +coordinates, then interpolation is required. + +Interpolation {#interp} +--------------------- + +Interpolation is the process of producing values at continuous coordinates from data sampled at discrete +coordinates. "Nearest-neighbor" and "N-Linear" are the two most commonly used interpolation methods. + + +Pixel coordinates {#pix-coords} +--------------------- + +**The pixel center is the origin of the continuous coordinate system.** + +### Top-left convention + +A common alternative convention is for the origin in the continuous space is at the "top-left" of the pixel. +This is not recommended, but can be acheived by explicitly adding a half-pixel translation, for example: + +```json +{ + "name": "center_to_top-left", + "type": "translation", + "translation" : [0.5, 0.5], + "output_space" : "top-left-space" +} +``` + +Coordinate Transformations {#coord-tforms} +===================== + +This document describes background and motivation that is outside the NGFF specification. + + +Direction {#direction} +--------------------- + +Specified coordinate transforms are in the "forward" direction. They represent functions +from *points* in the input space to *points* in the output space. For example, the transformation `"ij2xy"` + + +```json +{ + "name": "ij2xy", + "type": "scale", + "scale": [2, 0.5] + "input_axes" : ["i", "j"] + "output_axes" : ["x", "y"] +} +``` + +representes the function + +``` +x = 2 * i +y = 0.5 * j +``` + + +Recommendations {#recommendations} +===================== + + +"Native" physical space +--------------------- + +Datasets SHOULD define a transformation from array space to their "native physical space." +This transformation SHOULD describe physical pixel spacing and origin only, and therefore SHOULD consist of +`scale` and/or `translation` types only. + +Subsequent reorientation / registration transformations SHOULD use this native space as their `input_space`, +i.e., transformations should be defined in physical coordinates. + +