We'll be hosting office hours on Wednesdays, rotating between +APAC/AU/EU- and AMER/AU/EU-friendly times. + +Please join us if you have questions about NGFF and want to learn more about +the OME-Zarr storage format. We'll discuss the specification, the implementations, +what's new in the ecosystem, how you can get involved in the community and +much more.
+ +Please see the NGFF community calendar for exact +timings:
+ + + +Download the .ics file and add it to your calendar so won't miss any of our meetings! diff --git a/conf.py b/conf.py new file mode 100644 index 00000000..3b941302 --- /dev/null +++ b/conf.py @@ -0,0 +1,79 @@ +# Configuration file for the Sphinx documentation builder. +# +# For the full list of built-in configuration values, see the documentation: +# https://www.sphinx-doc.org/en/master/usage/configuration.html + +# -- Project information ----------------------------------------------------- +# https://www.sphinx-doc.org/en/master/usage/configuration.html#project-information + +project = 'NGFF' +copyright = '2023, NGFF Community' +author = 'NGFF Community' + +# -- General configuration --------------------------------------------------- +# https://www.sphinx-doc.org/en/master/usage/configuration.html#general-configuration + +extensions = ["myst_parser"] +source_suffix = [".rst", ".md"] + +templates_path = ['_templates'] +exclude_patterns = ['_build', 'Thumbs.db', '.DS_Store', '.git', '.pytest_cache', '**/.pytest_cache', '**/.tox', 'README.md', 'LICENSE.md', 'CONTRIBUTING.md'] + + + +# -- Options for HTML output ------------------------------------------------- +# https://www.sphinx-doc.org/en/master/usage/configuration.html#options-for-html-output + +html_theme = 'sphinx_book_theme' + +html_static_path = ['_static'] + +html_css_files = [ + 'https://cdn.datatables.net/v/dt/dt-1.11.5/datatables.min.css', +] + +html_js_files = [ + 'https://cdn.datatables.net/v/dt/dt-1.11.5/datatables.min.js', + 'main.js', +] + +html_extra_path = [ + '_bikeshed', +] + +# #################################### +# Run bikeshed build +# #################################### + +def bikeshed(): + + import glob + import os + import shutil + import subprocess + + for index_file in ["latest/index.bs"] + glob.glob("[0-9]*/index.bs"): + + output_file = index_file.replace("bs", "html") + output_dir = os.path.dirname(output_file) + target_dir = os.path.join("_bikeshed", output_dir) + + run_bikeshed = True + + # Give the loop a chance to skip files if no build is needed/requested + if "BIKESHED" not in os.environ and os.path.exists(output_file): + src_time = os.path.getmtime(index_file) + out_time = os.path.getmtime(output_file) + if src_time < out_time: + print(f"{index_file} unchanged") + run_bikeshed = False + + if run_bikeshed: + subprocess.check_call(f"bikeshed spec {index_file} {output_file}", shell=True) + + if os.path.exists(target_dir): + shutil.rmtree(target_dir) + shutil.copytree(output_dir, target_dir) + +bikeshed() +del bikeshed diff --git a/data/index.md b/data/index.md new file mode 100644 index 00000000..f4cdcec0 --- /dev/null +++ b/data/index.md @@ -0,0 +1,20 @@ +Data Resources +============== + +| Catalog | Hosting | Zarr Files | Size | +| ------------------------------------------------------------------------ | -----------------------------------------------------| ------------ | -------- | +| [BIA Samples](https://bit.ly/bia-ome-ngff-samples) | EBI | 90 | 200 GB | +| [Cell Painting Gallery](https://github.com/broadinstitute/cellpainting-gallery) | AWS Open Data Program | 136 | 20 TB | +| [CZB-Zebrahub](https://zebrahub.ds.czbiohub.org/imaging) | czbiohub | 5 | 1.2 TB | +| [DANDI](https://dandiarchive.org/dandiset/000108) ([identifiers.org][dandi2],[github][dandi3]) | AWS Open Data Program | 3914 | 355 TB | +| [Glencoe](https://glencoesoftware.com/ngff) | Glencoe Software, Inc. | 8 | 165 GB | +| [IDR Samples](https://idr.github.io/ome-ngff-samples/) | EBI | 88 | 3 TB | +| [MoBIE](https://mobie.github.io/specs/ngff.html) | EMBL-HD | 21 | 2 TB | +| [Neural Dynamics](https://registry.opendata.aws/allen-nd-open-data/) | AWS Open Data Program | 90 | 200 TB | +| [Sanger](https://www.sanger.ac.uk/project/ome-zarr/) | Sanger, UK | 10 | 1 TB | +| [SpatialData](https://github.com/scverse/spatialdata-notebooks/tree/main/datasets) | EMBL-HD | 10 | 25 GB | +| [SSBD](https://ssbd.riken.jp/ssbd-ome-ngff-samples) | SSBD | 12 | 196 GB | +| [webKnossos](https://zarr.webknossos.org) | scalableminds GmbH | 69 | 70 TB | + +[dandi2]: https://identifiers.org/DANDI:000108 +[dandi3]: https://github.com/dandisets/000108 diff --git a/index.html b/index.html deleted file mode 100644 index 74f803c4..00000000 --- a/index.html +++ /dev/null @@ -1,6 +0,0 @@ - - - - -If you are not redirected in five seconds, click here.
- diff --git a/index.rst b/index.rst new file mode 100644 index 00000000..ded27a68 --- /dev/null +++ b/index.rst @@ -0,0 +1,28 @@ +.. NGFF documentation master file, created by + sphinx-quickstart on Tue Mar 14 08:54:12 2023. + You can adapt this file completely to your liking, but it should at least + contain the root `toctree` directive. + +Next-generation file formats (NGFF) +=================================== + +OME-NGFF is an imaging format specification being developed by the bioimaging community to +address issues of scalability and interoperability. +Please see the :doc:`about/index` section for an introduction. +The OME-NGFF specification is detailed under :doc:`specifications/index`. +Various Image viewers and other software for working with NGFF data +are listed on the :doc:`tools/index` page. +Sample NGFF datasets provided by the community can be found under :doc:`data/index`. + +.. toctree:: + :maxdepth: 2 + + about/index + community/index + data/index + publications/index + specifications/index + tools/index + +.. raw:: html + diff --git a/latest/examples/bf2raw/.config.json b/latest/examples/bf2raw/.config.json new file mode 100644 index 00000000..2525328c --- /dev/null +++ b/latest/examples/bf2raw/.config.json @@ -0,0 +1,3 @@ +{ + "schema": "schemas/bf2raw.schema" +} diff --git a/latest/examples/bf2raw/image.json b/latest/examples/bf2raw/image.json new file mode 100644 index 00000000..c5eadb8a --- /dev/null +++ b/latest/examples/bf2raw/image.json @@ -0,0 +1,3 @@ +{ + "bioformats2raw.layout" : 3 +} \ No newline at end of file diff --git a/latest/examples/bf2raw/plate.json b/latest/examples/bf2raw/plate.json new file mode 100644 index 00000000..bd98a16e --- /dev/null +++ b/latest/examples/bf2raw/plate.json @@ -0,0 +1,22 @@ +{ + "bioformats2raw.layout" : 3, + "plate" : { + "columns" : [ { + "name" : "1" + } ], + "name" : "Plate Name 0", + "wells" : [ { + "path" : "A/1", + "rowIndex" : 0, + "columnIndex" : 0 + } ], + "field_count" : 1, + "rows" : [ { + "name" : "A" + } ], + "acquisitions" : [ { + "id" : 0 + } ], + "version" : "0.4" + } +} diff --git a/latest/examples/label_strict/colors_properties.json b/latest/examples/label_strict/colors_properties.json index 7f1d6f20..c96820e1 100644 --- a/latest/examples/label_strict/colors_properties.json +++ b/latest/examples/label_strict/colors_properties.json @@ -3,23 +3,25 @@ "version": "0.5-dev", "colors": [ { - "label-value": 1, - "rgba": [255, 255, 255, 255] + "label-value": 0, + "rgba": [0, 0, 128, 128] }, { - "label-value": 4, - "rgba": [0, 255, 255, 128] + "label-value": 1, + "rgba": [0, 128, 0, 128] } ], "properties": [ { - "label-value": 1, + "label-value": 0, "area (pixels)": 1200, - "class": "foo" + "class": "intercellular space" }, { - "label-value": 4, - "area (pixels)": 1650 + "label-value": 1, + "area (pixels)": 1650, + "class": "cell", + "cell type": "neuron" } ], "source": { diff --git a/latest/examples/multiscales_strict/multiscales_transformations.json b/latest/examples/multiscales_strict/multiscales_transformations.json index c99040c2..80dbedab 100644 --- a/latest/examples/multiscales_strict/multiscales_transformations.json +++ b/latest/examples/multiscales_strict/multiscales_transformations.json @@ -5,12 +5,12 @@ { "name": "y", "type": "space", - "units": "micrometer" + "unit": "micrometer" }, { "name": "x", "type": "space", - "units": "micrometer" + "unit": "micrometer" } ], "datasets": [ diff --git a/latest/examples/ome/.config.json b/latest/examples/ome/.config.json new file mode 100644 index 00000000..8a611ccf --- /dev/null +++ b/latest/examples/ome/.config.json @@ -0,0 +1,3 @@ +{ + "schema": "schemas/ome.schema" +} diff --git a/latest/examples/ome/series-2.json b/latest/examples/ome/series-2.json new file mode 100644 index 00000000..be7e9ed6 --- /dev/null +++ b/latest/examples/ome/series-2.json @@ -0,0 +1,3 @@ +{ + "series" : [ "0", "1" ] +} diff --git a/latest/header.include b/latest/header.include index 28305fe4..e98bf6fe 100644 --- a/latest/header.include +++ b/latest/header.include @@ -11,7 +11,7 @@ diff --git a/latest/index.bs b/latest/index.bs
index 0ec5b1f1..498bcac7 100644
--- a/latest/index.bs
+++ b/latest/index.bs
@@ -26,60 +26,6 @@ Status Text: will be provided between numbered versions. Data written with these
Status Text: (an "editor's draft") will not necessarily be supported.
-Introduction {#intro}
-=====================
-
-Bioimaging science is at a crossroads. Currently, the drive to acquire more,
-larger, preciser spatial measurements is unfortunately at odds with our ability
-to structure and share those measurements with others. During a global pandemic
-more than ever, we believe fervently that global, collaborative discovery as
-opposed to the post-publication, "data-on-request" mode of operation is the
-path forward. Bioimaging data should be shareable via open and commercial cloud
-resources without the need to download entire datasets.
-
-At the moment, that is not the norm. The plethora of data formats produced by
-imaging systems are ill-suited to remote sharing. Individual scientists
-typically lack the infrastructure they need to host these data themselves. When
-they acquire images from elsewhere, time-consuming translations and data
-cleaning are needed to interpret findings. Those same costs are multiplied when
-gathering data into online repositories where curator time can be the limiting
-factor before publication is possible. Without a common effort, each lab or
-resource is left building the tools they need and maintaining that
-infrastructure often without dedicated funding.
-
-This document defines a specification for bioimaging data to make it possible
-to enable the conversion of proprietary formats into a common, cloud-ready one.
-Such next-generation file formats layout data so that individual portions, or
-"chunks", of large data are reference-able eliminating the need to download
-entire datasets.
-
-
-Why "NGFF"? {#why-ngff}
--------------------------------------------------------------------------------------------------
-
-A short description of what is needed for an imaging format is "a hierarchy
-of n-dimensional (dense) arrays with metadata". This combination of features
-is certainly provided by HDF5
-from the HDF Group, which a number of
-bioimaging formats do use. HDF5 and other larger binary structures, however,
-are ill-suited for storage in the cloud where accessing individual chunks
-of data by name rather than seeking through a large file is at the heart of
-parallelization.
-
-As a result, a number of formats have been developed more recently which provide
-the basic data structure of an HDF5 file, but do so in a more cloud-friendly way.
-In the [PyData](https://pydata.org/) community, the Zarr [[zarr]] format was developed
-for easily storing collections of [NumPy](https://numpy.org/) arrays. In the
-[ImageJ](https://imagej.net/) community, N5 [[n5]] was developed to work around
-the limitations of HDF5 ("N5" was originally short for "Not-HDF5").
-Both of these formats permit storing individual chunks of data either locally in
-separate files or in cloud-based object stores as separate keys.
-
-A [current effort](https://zarr-specs.readthedocs.io/en/core-protocol-v3.0-dev/protocol/core/v3.0.html)
-is underway to unify the two similar specifications to provide a single binary
-specification. The editor's draft will soon be entering a [request for comments (RFC)](https://github.com/zarr-developers/zarr-specs/issues/101) phase with the goal of having a first version early in 2021. As that
-process comes to an end, this document will be updated.
-
OME-NGFF {#ome-ngff}
--------------------
@@ -101,6 +47,14 @@ The key words “MUST”, “MUST NOT”, “REQUIRED”, “SHALL”, “SHALL
“RECOMMENDED”, “MAY”, and “OPTIONAL” are to be interpreted as described in
[RFC 2119](https://tools.ietf.org/html/rfc2119).
+
diff --git a/latest/index.bs b/latest/index.bs
index 0ec5b1f1..498bcac7 100644
--- a/latest/index.bs
+++ b/latest/index.bs
@@ -26,60 +26,6 @@ Status Text: will be provided between numbered versions. Data written with these
Status Text: (an "editor's draft") will not necessarily be supported.
-Introduction {#intro}
-=====================
-
-Bioimaging science is at a crossroads. Currently, the drive to acquire more,
-larger, preciser spatial measurements is unfortunately at odds with our ability
-to structure and share those measurements with others. During a global pandemic
-more than ever, we believe fervently that global, collaborative discovery as
-opposed to the post-publication, "data-on-request" mode of operation is the
-path forward. Bioimaging data should be shareable via open and commercial cloud
-resources without the need to download entire datasets.
-
-At the moment, that is not the norm. The plethora of data formats produced by
-imaging systems are ill-suited to remote sharing. Individual scientists
-typically lack the infrastructure they need to host these data themselves. When
-they acquire images from elsewhere, time-consuming translations and data
-cleaning are needed to interpret findings. Those same costs are multiplied when
-gathering data into online repositories where curator time can be the limiting
-factor before publication is possible. Without a common effort, each lab or
-resource is left building the tools they need and maintaining that
-infrastructure often without dedicated funding.
-
-This document defines a specification for bioimaging data to make it possible
-to enable the conversion of proprietary formats into a common, cloud-ready one.
-Such next-generation file formats layout data so that individual portions, or
-"chunks", of large data are reference-able eliminating the need to download
-entire datasets.
-
-
-Why "NGFF"? {#why-ngff}
--------------------------------------------------------------------------------------------------
-
-A short description of what is needed for an imaging format is "a hierarchy
-of n-dimensional (dense) arrays with metadata". This combination of features
-is certainly provided by HDF5
-from the HDF Group, which a number of
-bioimaging formats do use. HDF5 and other larger binary structures, however,
-are ill-suited for storage in the cloud where accessing individual chunks
-of data by name rather than seeking through a large file is at the heart of
-parallelization.
-
-As a result, a number of formats have been developed more recently which provide
-the basic data structure of an HDF5 file, but do so in a more cloud-friendly way.
-In the [PyData](https://pydata.org/) community, the Zarr [[zarr]] format was developed
-for easily storing collections of [NumPy](https://numpy.org/) arrays. In the
-[ImageJ](https://imagej.net/) community, N5 [[n5]] was developed to work around
-the limitations of HDF5 ("N5" was originally short for "Not-HDF5").
-Both of these formats permit storing individual chunks of data either locally in
-separate files or in cloud-based object stores as separate keys.
-
-A [current effort](https://zarr-specs.readthedocs.io/en/core-protocol-v3.0-dev/protocol/core/v3.0.html)
-is underway to unify the two similar specifications to provide a single binary
-specification. The editor's draft will soon be entering a [request for comments (RFC)](https://github.com/zarr-developers/zarr-specs/issues/101) phase with the goal of having a first version early in 2021. As that
-process comes to an end, this document will be updated.
-
OME-NGFF {#ome-ngff}
--------------------
@@ -101,6 +47,14 @@ The key words “MUST”, “MUST NOT”, “REQUIRED”, “SHALL”, “SHALL
“RECOMMENDED”, “MAY”, and “OPTIONAL” are to be interpreted as described in
[RFC 2119](https://tools.ietf.org/html/rfc2119).
++Transitional metadata is added to the specification with the +intention of removing it in the future. Implementations may be expected (MUST) or +encouraged (SHOULD) to support the reading of the data, but writing will usually +be optional (MAY). Examples of transitional metadata include custom additions by +implementations that are later submitted as a formal specification. (See [[#bf2raw]]) +
+ Some of the JSON examples in this document include commments. However, these are only for clarity purposes and comments MUST NOT be included in JSON objects. @@ -139,7 +93,7 @@ For this example we assume an image with 5 dimensions and axes called `t,c,z,y,x ├── .zgroup # Each image is a Zarr group, or a folder, of other groups and arrays. ├── .zattrs # Group level attributes are stored in the .zattrs file and include │ # "multiscales" and "omero" (see below). In addition, the group level attributes - │ # must also contain "_ARRAY_DIMENSIONS" if this group directly contains multi-scale arrays. + │ # may also contain "_ARRAY_DIMENSIONS" for compatibility with xarray if this group directly contains multi-scale arrays. │ ├── 0 # Each multiscale level is stored as a separate Zarr array, │ ... # which is a folder containing chunk files which compose the array. @@ -242,9 +196,89 @@ keys as specified below for discovering certain types of data, especially images If part of [[#multiscale-md]], the length of "axes" MUST be equal to the number of dimensions of the arrays that contain the image data. +"bioformats2raw.layout" (transitional) {#bf2raw} +------------------------------------------------ + +[=Transitional=] "bioformats2raw.layout" metadata identifies a group which implicitly describes a series of images. +The need for the collection stems from the common "multi-image file" scenario in microscopy. Parsers like Bio-Formats +define a strict, stable ordering of the images in a single container that can be used to refer to them by other tools. + +In order to capture that information within an OME-NGFF dataset, `bioformats2raw` internally introduced a wrapping layer. +The bioformats2raw layout has been added to v0.4 as a transitional specification to specify filesets that already exist +in the wild. An upcoming NGFF specification will replace this layout with explicit metadata. + +Layout
+ +Typical Zarr layout produced by running `bioformats2raw` on a fileset that contains more than one image (series > 1): + ++series.ome.zarr # One converted fileset from bioformats2raw + ├── .zgroup + ├── .zattrs # Contains "bioformats2raw.layout" metadata + ├── OME # Special group for containing OME metadata + │ ├── .zgroup + │ ├── .zattrs # Contains "series" metadata + │ └── METADATA.ome.xml # OME-XML file stored within the Zarr fileset + ├── 0 # First image in the collection + ├── 1 # Second image in the collection + └── ... ++ +
Attributes
+ +The top-level `.zattrs` file must contain the `bioformats2raw.layout` key: ++path: examples/bf2raw/image.json +highlight: json ++ +If the top-level group represents a plate, the `bioformats2raw.layout` metadata will be present but +the "plate" key MUST also be present, takes precedence and parsing of such datasets should follow [[#plate-md]]. It is not +possible to mix collections of images with plates at present. + +
+path: examples/bf2raw/plate.json +highlight: json ++ +The `.zattrs` file within the OME group may contain the "series" key: + +
+path: examples/ome/series-2.json +highlight: json ++ +
Details
+ +Conforming groups: + +- MUST have the value "3" for the "bioformats2raw.layout" key in their `.zattrs` metadata at the top of the hierarchy; +- SHOULD have OME metadata representing the entire collection of images in a file named "OME/METADATA.ome.xml" which: + - MUST adhere to the OME-XML specification but + - MUST use `path: examples/label_strict/colors_properties.json highlight: json+In this case, the pixels consisting of a 0 in the Zarr array will be displayed as 50% blue and 50% opacity. Pixels with a 1 in the Zarr array, +which correspond to cellular space, will be displayed as 50% green and 50% opacity. + "plate" metadata {#plate-md} ---------------------------- @@ -452,7 +492,7 @@ contain only alphanumeric characters, MUST be case-sensitive, and MUST NOT be a other `name` in the `rows` list. Care SHOULD be taken to avoid collisions on case-insensitive filesystems (e.g. avoid using both `Aa` and `aA`). -The `plate` dictionary SHOULD contain a `version` key whose value MUST be a string specifying the +The `plate` dictionary MUST contain a `version` key whose value MUST be a string specifying the version of the plate specification. The `plate` dictionary MUST contain a `wells` key whose value MUST be a list of JSON objects @@ -552,10 +592,12 @@ Projects which support reading and/or writing OME-NGFF data include:
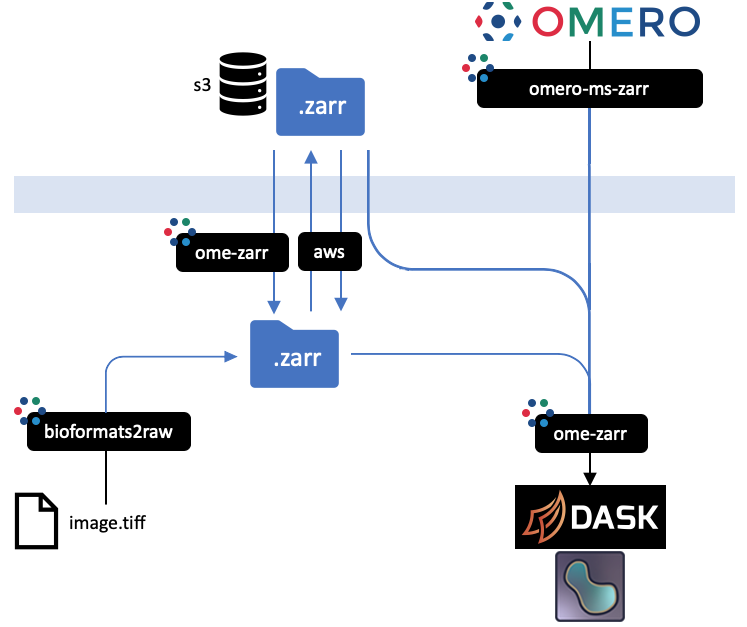 +
+ All implementations prevent an equivalent representation of a dataset which can be downloaded or uploaded freely. An interactive
version of this diagram is available from the [OME2020 Workshop](https://downloads.openmicroscopy.org/presentations/2020/Dundee/Workshops/NGFF/zarr_diagram/).
@@ -583,6 +625,16 @@ Version History {#history}
All implementations prevent an equivalent representation of a dataset which can be downloaded or uploaded freely. An interactive
version of this diagram is available from the [OME2020 Workshop](https://downloads.openmicroscopy.org/presentations/2020/Dundee/Workshops/NGFF/zarr_diagram/).
@@ -583,6 +625,16 @@ Version History {#history}