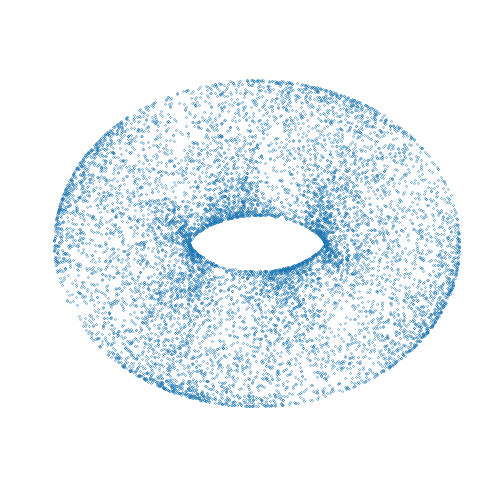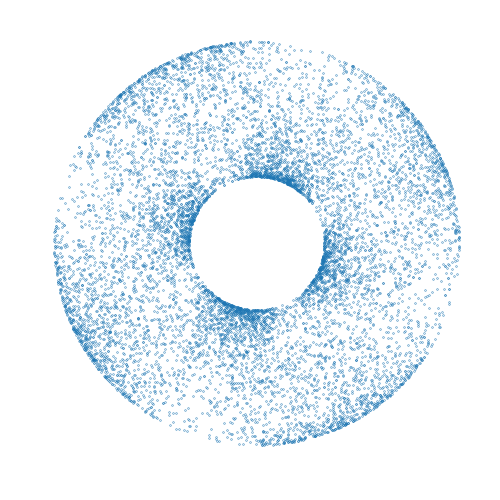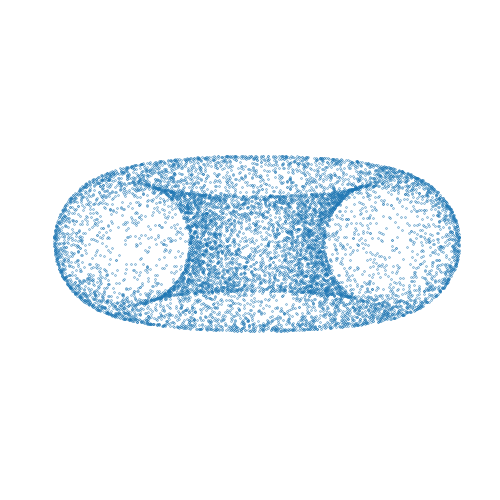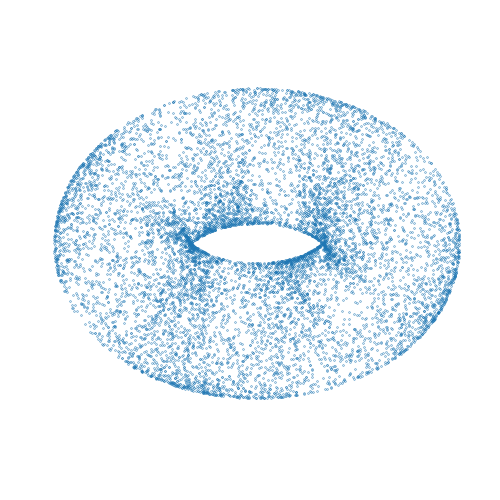
Mici is a Python package providing implementations of Markov chain Monte Carlo (MCMC) methods for approximate inference in probabilistic models, with a particular focus on MCMC methods based on simulating Hamiltonian dynamics on a manifold.
Key features include
- a modular design allowing use of a wide range of inference algorithms by mixing and matching different components, and making it easy to extend the package,
- a pure Python code base with minimal dependencies, allowing easy integration within other code,
- built-in support for several automatic differentiation frameworks, including JAX and Autograd, or the option to supply your own derivative functions,
- implementations of MCMC methods for sampling from distributions on embedded manifolds implicitly-defined by a constraint equation and distributions on Riemannian manifolds with a user-specified metric,
- computationally efficient inference via transparent caching of the results of expensive operations and intermediate results calculated in derivative computations allowing later reuse without recalculation,
- memory efficient inference for large models by memory-mapping chains to disk, allowing long runs on large models without hitting memory issues.
To install and use Mici the minimal requirements are a Python 3.10+ environment with NumPy and SciPy installed. The latest Mici release on PyPI (and its dependencies) can be installed in the current Python environment by running
pip install miciTo instead install the latest development version from the main branch on Github run
pip install git+https://github.com/matt-graham/miciIf available in the installed Python environment the following additional packages provide extra functionality and features
- ArviZ: if ArviZ is
available the traces (dictionary) output of a sampling run can be directly
converted to an
arviz.InferenceDatacontainer object usingarviz.convert_to_inference_dataor implicitly converted by passing the traces dictionary as thedataargument to ArviZ API functions, allowing straightforward use of the ArviZ's extensive visualisation and diagnostic functions. - Autograd: if available Autograd will
be used to automatically compute the required derivatives of the model
functions (providing they are specified using functions from the
autograd.numpyandautograd.scipyinterfaces). To sample chains in parallel usingautogradfunctions you also need to install multiprocess. This will causemultiprocess.Poolto be used in preference to the in-builtmutiprocessing.Poolfor parallelisation as multiprocess supports serialisation (via dill) of a much wider range of types, including of Autograd generated functions. Both Autograd and multiprocess can be installed alongside Mici by runningpip install mici[autograd]. - JAX: if available JAX will be used to
automatically compute the required derivatives of the model functions (providing
they are specified using functions from the
jaxinterface). To sample chains parallel using JAX functions you also need to install multiprocess, though note due to JAX's use of multithreading which is incompatible with forking child processes, this can result in deadlock. Both JAX and multiprocess can be installed alongside Mici by runningpip install mici[jax]. - SymNum: if available SymNum will be used to
automatically compute the required derivatives of the model functions (providing
they are specified using functions from the
symnum.numpyinterface). Symnum can be installed alongside Mici by runningpip install mici[symnum].
Mici is named for Augusta 'Mici' Teller, who along with Arianna Rosenbluth developed the code for the MANIAC I computer used in the seminal paper Equations of State Calculations by Fast Computing Machines which introduced the first example of a Markov chain Monte Carlo method.
Other Python packages for performing MCMC inference include PyMC, PyStan (the Python interface to Stan), Pyro / NumPyro, TensorFlow Probability, emcee, Sampyl and BlackJAX.
Unlike PyMC, PyStan, (Num)Pyro and TensorFlow Probability which are complete probabilistic programming frameworks including functionality for defining a probabilistic model / program, but like emcee, Sampyl and BlackJAX, Mici is solely focused on providing implementations of inference algorithms, with the user expected to be able to define at a minimum a function specifying the negative log (unnormalized) density of the distribution of interest.
Further while PyStan, (Num)Pyro and TensorFlow Probability all push the sampling loop into external compiled non-Python code, in Mici the sampling loop is run directly within Python. This has the consequence that for small models in which the negative log density of the target distribution and other model functions are cheap to evaluate, the interpreter overhead in iterating over the chains in Python can dominate the computational cost, making sampling much slower than packages which outsource the sampling loop to a efficient compiled implementation.
API documentation for the package is available
here. The three main user-facing
modules within the mici package are the systems, integrators and
samplers modules and you will generally need to create an instance of one
class from each module.
mici.systems -
Hamiltonian systems encapsulating model functions and their derivatives
EuclideanMetricSystem- systems with a metric on the position space with a constant matrix representation,GaussianEuclideanMetricSystem- systems in which the target distribution is defined by a density with respect to the standard Gaussian measure on the position space allowing analytically solving for flow corresponding to the quadratic components of Hamiltonian (Shahbaba et al., 2014),RiemannianMetricSystem- systems with a metric on the position space with a position-dependent matrix representation (Girolami and Calderhead, 2011),SoftAbsRiemannianMetricSystem- system with SoftAbs eigenvalue-regularized Hessian of negative log target density as metric matrix representation (Betancourt, 2013),DenseConstrainedEuclideanMetricSystem- Euclidean-metric system subject to holonomic constraints (Hartmann and Schütte, 2005; Brubaker, Salzmann and Urtasun, 2012; Lelièvre, Rousset and Stoltz, 2019) with a dense constraint function Jacobian matrix,
mici.integrators -
symplectic integrators for Hamiltonian dynamics
LeapfrogIntegrator- explicit leapfrog (Störmer-Verlet) integrator for separable Hamiltonian systems (Leimkulher and Reich, 2004),ImplicitLeapfrogIntegrator- implicit (or generalized) leapfrog integrator for Hamiltonian systems with non-separable component (Leimkulher and Reich, 2004),ImplicitMidpointIntegrator- implicit midpoint integrator for general Hamiltonian systems (Leimkulher and Reich, 2004),SymmetricCompositionIntegrator- family of symplectic integrators for Hamiltonians that can be split in to components with tractable flow maps, with specific two-, three- and four-stage instantations due to Blanes, Casas and Sanz-Serna (2014),ConstrainedLeapfrogIntegrator- constrained leapfrog integrator for Hamiltonian systems subject to holonomic constraints (Andersen, 1983; Leimkuhler and Reich, 1994).
mici.samplers - MCMC
samplers for peforming inference
StaticMetropolisHMC- static integration time Hamiltonian Monte Carlo with Metropolis accept step (Duane et al., 1987),RandomMetropolisHMC- random integration time Hamiltonian Monte Carlo with Metropolis accept step (Mackenzie, 1989),DynamicSliceHMC- dynamic integration time Hamiltonian Monte Carlo with slice sampling from trajectory, equivalent to the original 'NUTS' algorithm (Hoffman and Gelman, 2014).DynamicMultinomialHMC- dynamic integration time Hamiltonian Monte Carlo with multinomial sampling from trajectory, equivalent to the current default MCMC algorithm in Stan (Hoffman and Gelman, 2014; Betancourt, 2017).
The manifold MCMC methods implemented in Mici have been used in several research projects. Below links are provided to a selection of Jupyter notebooks associated with these projects as demonstrations of how to use Mici and to illustrate some of the settings in which manifold MCMC methods can be computationally advantageous.
| Manifold lifting: scaling MCMC to the vanishing noise regime | |
|---|---|
| Open non-interactive version with nbviewer |
|
| Open interactive version with Binder |
|
| Open interactive version with Google Colab |
|
| Manifold MCMC methods for inference in diffusion models | |
|---|---|
| Open non-interactive version with nbviewer |
|
| Open interactive version with Binder |
|
| Open interactive version with Google Colab |
|
A simple complete example of using the package to compute approximate samples from a
distribution on a two-dimensional torus embedded in a three-dimensional space is given
below. The computed samples are visualized in the animation above. Here we use
SymNum to automatically construct functions to
calculate the required derivatives (gradient of negative log density of target
distribution and Jacobian of constraint function), sample four chains in parallel using
multiprocessing, use ArviZ to calculate
diagnostics and use Matplotlib to plot the samples.
import mici
import numpy as np
import symnum
import symnum.numpy as snp
import matplotlib.pyplot as plt
import matplotlib.animation as animation
import arviz
# Define fixed model parameters
R = 1.0 # toroidal radius ∈ (0, ∞)
r = 0.5 # poloidal radius ∈ (0, R)
α = 0.9 # density fluctuation amplitude ∈ [0, 1)
# State dimension
dim_q = 3
# Define constraint function such that the set {q : constr(q) == 0} is a torus
@symnum.numpify(dim_q)
def constr(q):
x, y, z = q
return snp.array([((x**2 + y**2) ** 0.5 - R) ** 2 + z**2 - r**2])
# Define negative log density for the target distribution on torus
# (with respect to 2D 'area' measure for torus)
@symnum.numpify(dim_q)
def neg_log_dens(q):
x, y, z = q
θ = snp.arctan2(y, x)
ϕ = snp.arctan2(z, x / snp.cos(θ) - R)
return snp.log1p(r * snp.cos(ϕ) / R) - snp.log1p(snp.sin(4 * θ) * snp.cos(ϕ) * α)
# Specify constrained Hamiltonian system with default identity metric
system = mici.systems.DenseConstrainedEuclideanMetricSystem(
neg_log_dens,
constr,
backend="symnum",
)
# System is constrained therefore use constrained leapfrog integrator
integrator = mici.integrators.ConstrainedLeapfrogIntegrator(system)
# Seed a random number generator
rng = np.random.default_rng(seed=1234)
# Use dynamic integration-time HMC implementation as MCMC sampler
sampler = mici.samplers.DynamicMultinomialHMC(system, integrator, rng)
# Sample initial positions on torus using parameterisation (θ, ϕ) ∈ [0, 2π)²
# x, y, z = (R + r * cos(ϕ)) * cos(θ), (R + r * cos(ϕ)) * sin(θ), r * sin(ϕ)
n_chain = 4
θ_init, ϕ_init = rng.uniform(0, 2 * np.pi, size=(2, n_chain))
q_init = np.stack(
[
(R + r * np.cos(ϕ_init)) * np.cos(θ_init),
(R + r * np.cos(ϕ_init)) * np.sin(θ_init),
r * np.sin(ϕ_init),
],
-1,
)
# Define function to extract variables to trace during sampling
def trace_func(state):
x, y, z = state.pos
return {"x": x, "y": y, "z": z}
# Sample 4 chains in parallel with 500 adaptive warm up iterations in which the
# integrator step size is tuned, followed by 2000 non-adaptive iterations
final_states, traces, stats = sampler.sample_chains(
n_warm_up_iter=500,
n_main_iter=2000,
init_states=q_init,
n_process=4,
trace_funcs=[trace_func],
)
# Print average accept probability and number of integrator steps per chain
for c in range(n_chain):
print(f"Chain {c}:")
print(f" Average accept prob. = {stats['accept_stat'][c].mean():.2f}")
print(f" Average number steps = {stats['n_step'][c].mean():.1f}")
# Print summary statistics and diagnostics computed using ArviZ
print(arviz.summary(traces))
# Visualize concatentated chain samples as animated 3D scatter plot
fig, ax = plt.subplots(
figsize=(4, 4),
subplot_kw={"projection": "3d", "proj_type": "ortho"},
)
(points_3d,) = ax.plot(*(np.concatenate(traces[k]) for k in "xyz"), ".", ms=0.5)
ax.axis("off")
for set_lim in [ax.set_xlim, ax.set_ylim, ax.set_zlim]:
set_lim((-1, 1))
def update(i):
angle = 45 * (np.sin(2 * np.pi * i / 60) + 1)
ax.view_init(elev=angle, azim=angle)
return (points_3d,)
anim = animation.FuncAnimation(fig, update, frames=60, interval=100)
plt.show()- Andersen, H.C., 1983. RATTLE: A “velocity”
version of the SHAKE algorithm for molecular dynamics calculations.
Journal of Computational Physics, 52(1), pp.24-34.
- Duane, S., Kennedy, A.D., Pendleton, B.J. and
Roweth, D., 1987. Hybrid Monte Carlo. Physics letters B, 195(2),
pp.216-222.
- Mackenzie, P.B., 1989. An improved
hybrid Monte Carlo method. Physics Letters B, 226(3-4), pp.369-371.
- Horowitz, A.M., 1991. A generalized
guided Monte Carlo algorithm. Physics Letters B, 268(CERN-TH-6172-91),
pp.247-252.
- Leimkuhler, B. and Reich, S., 1994.
Symplectic integration of constrained Hamiltonian systems. Mathematics of
Computation, 63(208), pp.589-605.
- Leimkuhler, B. and Reich, S., 2004.
Simulating Hamiltonian dynamics (Vol. 14). Cambridge University Press.
- Hartmann, C. and Schütte, C., 2005. A
constrained hybrid Monte‐Carlo algorithm and the problem of calculating the
free energy in several variables. ZAMM ‐ Journal of Applied Mathematics and
Mechanics, 85(10), pp.700-710.
- Girolami, M. and Calderhead, B., 2011.
Riemann manifold Langevin and Hamiltonian Monte Carlo methods. Journal of
the Royal Statistical Society: Series B (Statistical Methodology), 73(2), pp.123-214.
- Brubaker, M., Salzmann, M. and
Urtasun, R., 2012. A family of MCMC methods on implicitly defined
manifolds. In Artificial intelligence and statistics (pp. 161-172).
- Betancourt, M., 2013. A general metric
for Riemannian manifold Hamiltonian Monte Carlo. In Geometric science of
information (pp. 327-334).
- Hoffman, M.D. and Gelman, A., 2014. The
No-U-turn sampler: adaptively setting path lengths in Hamiltonian Monte
Carlo. Journal of Machine Learning Research, 15(1), pp.1593-1623.
- Shahbaba, B., Lan, S., Johnson, W.O. and
Neal, R.M., 2014. Split Hamiltonian Monte Carlo. Statistics and
Computing, 24(3), pp.339-349.
- Blanes, S., Casas, F., & Sanz-Serna, J. M., 2014.
Numerical integrators for the Hybrid Monte Carlo method.
SIAM Journal on Scientific Computing, 36(4), A1556-A1580.
- Betancourt, M., 2017. A conceptual
introduction to Hamiltonian Monte Carlo.
- Lelièvre, T., Rousset, M. and Stoltz, G., 2019. Hybrid Monte Carlo methods for sampling probability measures on
submanifolds. In Numerische Mathematik, 143(2), (pp.379-421).